10 Building R Packages
“There are two ways of constructing a software design: One way is to make it so simple that there are obviously no deficiencies, and the other way is to make it so complicated that there are no obvious deficiencies. The first method is far more difficult.”
- Tony Hoare, Pioneering British computer scientist
10.1 Introduction
One of strengths of R is its capacity to format and share user-designed software as packages. Clearly it is possible to apply R for one’s entire scientific career without creating an R package. However, development of a package, even if it is not distributed to a formal repository, ensures that your software is trustworthy and portable. Importantly, this chapter only provides a overview of basic topics in package development. The most thorough and up-to-date guide to package creation is the document Writing R Extensions, which is maintained by the the R development core team. This chapter will require some knowledge of system shell commands, as described in Section 9.2.
10.2 Package Components
An R package is a directory of files, generally with nested subdirectories. Specifically,
-
DESCRIPTIONandNAMESPACEfiles define fundamental characteristics of the package, e.g., the author(s), the maintainer, the package version, the dependency on other packages, etc. - Package subdirectories, and their nested files, contain the package contents. The following subdirectories are possible, although not all need to exist within a package.
- The
Rsubdirectory contains the package R code, stored as .r files, and will almost always exist in R packages. - The optional
datasubdirectory contains package datasets, usually stored as .rda files, which can be created usingsave(). - The
mansubdirectory contains the reuiqred package documentation, stored as .rd files, for functions (in theRdirectory) and data (indata), and thus, will always exist in packages. - The optional
srcsubdirectory contains raw source code requiring compilation (C, C++, Fortran). When building a package R will callR CMD SHLIB(see Section 9.3) to create appropriate binary shared library files. - Other potential subdirectories include:
demo,exec,inst,po,tests,tools, andvignettes.
- The
Fig 10.1 shows the contents of the R package streamDAG (K. Aho, Kriloff, et al. 2023). These directories, and their files, are contained within a parent directory called streamDAG.

FIGURE 10.1: Subdirectory level components of the streamDAG package.
Example 10.1 \(\text{}\)
Creation of package components can be facilitated with the function package.skeleton() . From the package.skeleton() documentation Examples (see ?package.skeleton), assume that we want to build a package that contains two silly functions: (f and g) and two silly datasets: (d and e).
f <- function(x, y) x + y
g <- function(x, y) x - y
d <- data.frame(a = 1, b = 2)
e <- rnorm(1000)We specify these as the list argument in package.skeleton() and give the package the name mypkg.
package.skeleton(list = c("f","g","d","e"), name = "mypkg")Creating directories ...
Creating DESCRIPTION ...
Creating NAMESPACE ...
Creating Read-and-delete-me ...
Saving functions and data ...
Making help files ...
Done.
Further steps are described in './mypkg/Read-and-delete-me'.Running this code will cause a package skeleton for mypkg to be generated and sent to the working directory. Note that the skeleton contains the subdirectories: data, r, and man. The datasets d and e were converted to .rda files by package.skeleton() and were placed in the data subdirectory. The functions f and g were converted to .r files and placed in the r subdirectory. Documentation skeletons for both functions and both datasets, as .rd files, were placed in the man subdirectory. Package DESCRIPTION, NAMESPACE files, and a throw-away (Read-and-delete-me) file were also created. This structure is revealed by the BASH
tree utility (Table 9.1). Here I open the Ubuntu version of Linux implemented in WSL from cmd.
Here I navigate to the mypkg parent directory, and implement tree.
$ cd /mnt/c/Users/ahoken/Documents/GitHub/Amalgam/mypkg
/mnt/c/Users/ahoken/Documents/GitHub/Amalgam/mypkg$ tree .
├── DESCRIPTION
├── NAMESPACE
├── R
│ ├── f.R
│ └── g.R
├── Read-and-delete-me
├── data
│ ├── d.rda
│ └── e.rda
└── man
├── d.Rd
├── e.Rd
├── f.Rd
├── g.Rd
└── mypkg-package.Rd\(\blacksquare\)
Example 10.2 \(\text{}\)
This chapter will largely follow the development of a custom package based on functions and methods for alpha diversity created in Example 8.22 from Chapter 8. Recall that Example 8.22 resulted in three functions: alpha.div(), print.a_div(), and plot.a_div(). The latter two functions were S3 printing and plotting methods for objects of class a_div, resulting from application of alpha_div().
I will develop the package within this book’s working directory. As a first step, I will create a directory named mydiv to house the package contents. Within this directory, I create r and man subdirectories, and skeleton DESCRIPTION, NAMESPACE text files (see Sections 10.6 and 10.7). I also place the functions alpha.div(), plot.a_div(), and plot.a_div() in an r directory, and place a skeleton documentation file for the functions (see Example 10.3) in the man subdirectory.
.
├── DESCRIPTION
├── NAMESPACE
├── R
│ ├── alpha_div.R
│ ├── plot.a_div.R
│ └── print.a_div.R
└── man
└── alpha_div.RdThe entire package directory system is available here.
\(\blacksquare\)
10.3 Datasets (the data Subdirectory)
Datasets in R are stored in the data subdirectory. Three data formats are possible:
- Raw .r code
- Tabular data (e.g., .txt, .csv files)
- Data “images” created using the function save(), e.g., .rda or .Rdata files. This approach is generally recommended, particularly for large datasets. Here we create a simple .rda dataset, and send it to the working directory.
Data from packages will either be accessible via lazy loading (which allows increased accessibility) or with the data() function (see Sections 8.8.1.1.2 and 8.8.2). Under the former approach, package data objects will not be loaded upon loading of their package environment, however promises are created, requiring the object to be loaded when its name is entered in a session. Lazy loading always occurs for package R code but is optional for package data. Lazy loading of data can be specified in a ‘LazyData’ field from a package’s DESCRIPTION file (see below). Examples of lazy loaded data include objects from the package datasets. Note that these do not require data() for loading:
# data describing Biochemical Oxygen Demand
datasets::BOD Time demand
1 1 8.3
2 2 10.3
3 3 19.0
4 4 16.0
5 5 15.6
6 7 19.8Under the latter, more common approach, data(foo) must be called to allow availability of the dataset foo.
resources avail y1 n1
1 Riparian 0.06 0 445
2 Conifer 0.13 6 445
3 Mt. Shrub 1 0.16 9 445
4 Aspen 0.15 18 445
5 Rock outcrop 0.06 14 445
6 Sage/Bitterbrush 0.17 63 445
7 Windblown ridges 0.12 46 445
8 Mt shrub 2 0.04 62 445
9 Prescribed burns 0.09 178 445
10 Clearcut 0.02 49 445Note that the mydiv package (Example 10.2) does not contain data, and does not have a data directory.
10.4 R Code (the r Subdirectory)
Code for functions is generally stored in the r directory, as .r files. IDEs like RStudio, which allows straightforward generation of .r scripts, e.g., File > New File > R script, can greatly aid in this process. Single .r files can contain multiple functions, although a one function per file approach may be easier to manage. Recall that individual .r files were created for alpha.div and its methods, in mydiv package (Example 10.2).
10.5 Documentation (the man Subdirectory)
As functions become complex, it may become difficult to keep track of the meaning of function arguments, and the characteristics of function output, using a simple notes-to-self approach, e.g., . R documentation (.rd) files provide a framework for documenting, R functions, methods, and datasets. The prompt() family of functions can greatly facilitate the creation of .rd files. In Example 10.1, the function package.skelton() used the functions prompt() and promptData() to build documentation skeletons for functions and datasets, respectively.
Example 10.3 \(\text{}\)
For instance, to build skeleton documentation file for the function alpha_div() (Example 10.2), I could use:
Created file named 'alpha_div.rd'.
Edit the file and move it to the appropriate directory.The file alpha_div.rd is generated, and sent to the working directory for further editing (Fig 10.2).
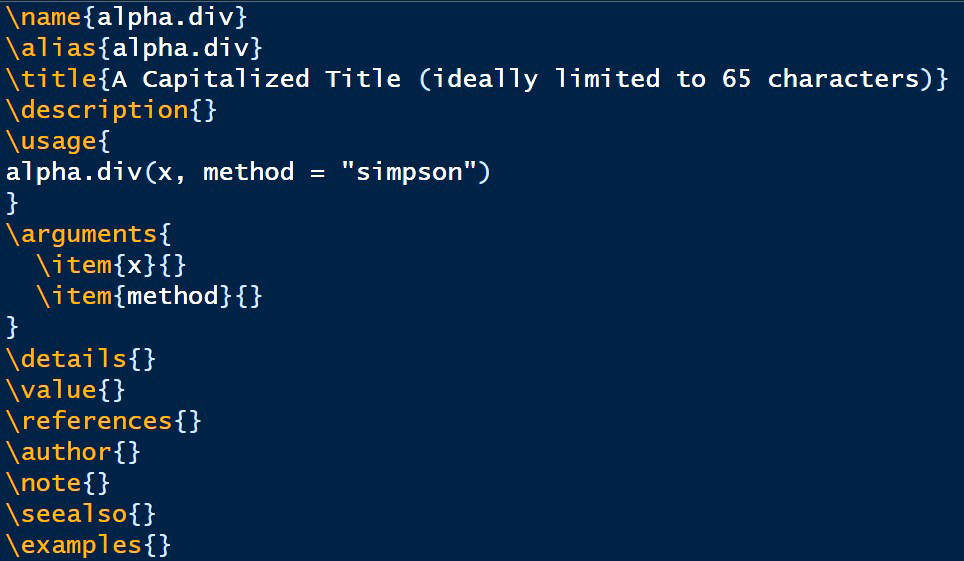
FIGURE 10.2: Documentation file skeleton for the function alpha.div(). Some auto-generated content omitted for brevity.
A completed version of alpha_div.rd (with empty documentation components filled in) can be found here.
A Preview widget will be available in the RStudio Code Editor pane (Section 2.10) when an .rd document is open:
![]() .
.
Clicking the widget will allow one to view the documentation file as a user would see it when requesting that documentation136.
Some guidance for completing .rd files is provided by notes in the skeleton generated by prompt() itself. I have removed these notes in Fig 10.2 to save space. As before, the authoritative resource for documentation building is Writing R Extensions.
\(\blacksquare\)
Package .rd files should be placed into the man directory. These files will be rendered into a single documentation entity (generally HTML or PDF) as the package is compiled. A single .rd file can be converted to legible documentation in HTML, PDF, or other formats, by using appropriate R CMD routines from a system shell (see Section 9.2). Important R CMD documentation rendering algorithms include:
-
R CMD Rd2pdf foo.rd, which will renderfoo.rdinto a PDF document. -
R CMD Rd2txt foo.rd, which will renderfoo.rdinto a pretty text format. -
R CMD Rdconv foo.rd, which can renderfoo.rdinto a variety of formats including plain text, HTML, or PDF.
Example 10.4 \(\text{}\)
Here I generate a PDF rendered version of alpha_div.Rd, by applying R CMD Rd2pdf after navigating to the mydiv man directory.
I get the following output:
Converting Rd files to LaTeX ...
alpha_div.Rd
Creating pdf output from LaTeX ...
texify: security risk: running with elevated privileges
Saving output to 'alpha_div.pdf' ...
Done
The rendered PDF can be obtained here.
\(\blacksquare\)
10.6 The DESCRIPTION File
The DESCRIPTION file contains basic information about a package. The DESCRIPTION file skeleton created by package.skeleton() has the form:
Package: mydiv
Type: Package
Title: What the package does (short line)
Version: 1.0
Date: 2026-1-23
Author: Who wrote it
Maintainer: Who to complain to <yourfault@somewhere.net>
Description: More about what it does (maybe more than one line)
License: What license is it under?The DESCRIPTION file will have a Debian control file format (see ?read.dcf. Specifically, fields in DESCRIPTION must start with the field name, comprised of ASCII (Ch 12) printable characters, followed by a colon. The value for the field is given after the colon and an additional space. If allowed, field values longer than one line must use a space or a tab to start a new line. Specification of ‘Package’, ‘Version’, ‘License’, ‘Description’, ‘Title’, ‘Author’, and ‘Maintainer’ fields, as shown above, are mandatory.
- The
‘Package’field gives the name of the package. - The
‘Version’field gives a user-specified package version. It should be a sequence of at least two non-negative integers separated by single usages‘.’and/or‘-’characters. - The
‘Title’field should provide a descriptive title for the package. It should use title case (capitals for principal words), and not have any continuation lines. - The
‘Author’field describes who wrote the package. Note that if your package contains wrappers of the work of others, which are included in thesrcdirectory, then you are not the sole author. - The
‘Maintainer’field provides a single name followed by a valid email address in angle brackets (see chunk above). - The
‘Description’field should provide a comprehensive description of what the package does. Several (complete) sentences, complete, although these should limited to one paragraph. The field value should not to start with the package name, or‘This package...’. - The
‘License’field provides standard open source license information for the package. Failure to specify license information may prevent others from legally using, or distributing your package. Standard licenses available from (https://www.R-project.org/Licenses/) include GPL-2, GPL-3, LGPL-2, LGPL-2.1, LGPL-3, AGPL-3, Artistic-2.0, BSD_2_clause, and BSD_3_clause MIT. See Writing R Extensions for more information. - Other optional fields include:
‘Copyright’,‘Date’,‘Depends’,‘Imports’,‘Suggests’,‘Enhances’,‘LinkingTo’,‘Additional_repositories’,‘SystemRequirements’,‘URL’,‘BugReports’,‘Collate’,‘LazyData’,‘KeepSource’,‘ByteCompile’,‘UseLTO’,‘StagedInstall’,‘Biarch’,‘BuildVignettes’,‘VignetteBuilder’,‘NeedsCompilation’,‘OS_type’, and‘Type’. See Writing R Extensions for more information on these fields.
Here is “finished” form of the mydiv DESCRIPTION file:
10.7 The NAMESPACE File
The R namespace management system allows package authors to specify which variables in the package can be exported to package users, and which variables should be imported from other packages. See Sections 8.8.1.1.2, 8.8.3 for additional information.
The mandatory NAMESPACE file for the toy mydiv package is extremely simple:
exportPattern(.)
importFrom("utils", "stack")
import(ggplot2)
S3method(print, a_div)
S3method(plot, a_div)- On Line 1, the file indicates that all objects in the package, and their associated names, can be exported using
exportPattern(.). - Lines 2 and 3 define external packages required by mydiv functions. In particular, import of exported variables from other packages requires specification of
importandimportFrom. Theimportdirective imports all exported variables from specified package(s). Thus,import(foo)imports all exported variables in the package foo. If a package requires some of the exported variables from a package, thenimportFromcan be used. TheNAMESPACEdirectiveimportFrom(foo, f, g)indicates thatfandgfrom package foo should be imported. - To ensure that S3 methods for package classes are available, one must register them in the
NAMESPACEfile (Lines 5-6). For instance, if a package has a functionprint.foo()that serves as a print method for classfoo, then one should includeS3method(print, foo)as a line inNAMESPACE.
10.8 Package Compilation
As with compilation of shared libraries from C and Fortran files using R CMD SHLIB (Section 9.3), and the rendering of individual .rd files (Example 10.4), the building and installation of a user-designed package from a system shell will depend on existing Environmental Paths on your machine (see Section 9.4.1). In the worst case, this will require depositing the package contents in the R directory containing the R CMD routines (currently C:\Program Files\R\R-4.5.2\bin\x64 on my Windows machine)137, and running R CMD routines. R CMD processes for package building138 include:
-
R CMD buildfoo, which would build the package foo. -
R CMD checkfoo.tar.gz, which would check the tarballed package foo.tar.gz, created byR CMD build. -
R CMD INSTALLfoo.tar.gz can be used to install the package foo.
Example 10.5 \(\text{}\)
Continuing from Example 10.1, I complete the following steps for package building/compression, checking, and installation.
- Here I navigate to the book’s home directory (which contains mydiv) and build a tarballed version of the package using:
R CMD build mydiv.
The following output is produced:
* checking for file 'mydiv/DESCRIPTION' ... OK
* preparing 'mydiv':
* checking DESCRIPTION meta-information ... OK
* checking for LF line-endings in source and make files and shell scripts
* checking for empty or unneeded directories
* building 'mydiv_1.0.tar.gz'- Here I “check” the functionality of the tarballed version of the package using:
R CMD check mypkg_1.0.tar.gz.
I get the following output (many lines omitted for brevity):
* using log directory 'C:/Users/ahoken/Documents/GitHub/Amalgam/mydiv.Rcheck'
* using R version 4.5.1 (2025-06-13 ucrt)
* using platform: x86_64-w64-mingw32
* R was compiled by
gcc.exe (GCC) 14.2.0
GNU Fortran (GCC) 14.2.0
* running under: Windows 11 x64 (build 26100)
* using session charset: UTF-8
* checking for file 'mydiv/DESCRIPTION' ... OK
* checking extension type ... Package
* this is package 'mydiv' version '1.0'
* checking package namespace information ... OK
* checking for missing documentation entries ... OK
* checking for code/documentation mismatches ... OK
* checking PDF version of manual ... OK
* DONEChecks are even more thorough if one uses the R CMD check option --as-cran, which performs assessments one must pass for submission to CRAN.
- Finally, I Install the mydiv package onto my workstation using:
R CMD INSTALL mydiv_1.0.tar.gz.
I get the following output:
* installing to library 'C:/Users/ahoken/AppData/Local/R/win-library/4.5'
* installing *source* package 'mydiv' ...
** this is package 'mydiv' version '1.0'
** using staged installation
** R
** byte-compile and prepare package for lazy loading
** help
*** installing help indices
** building package indices
** testing if installed package can be loaded from temporary location
** testing if installed package can be loaded from final location
** testing if installed package keeps a record of temporary installation path
* DONE (mydiv)\(\blacksquare\)
Exercises
Create an .rd documentation file for the function for McIntosh’s index of site biodiversity from Exercise 2 in 8. Make a .pdf or .html from the .rd file using the appropriate
R CMDroutines.-
Create an R package consisting of at least one function. Specifically,
- Create a skeleton of the package using
package.skeleton(). - Finish the .rd file(s) in
man. - Complete the
DESCRIPTIONfile. - Complete the
NAMESPACEfile. - Build the package using
R CMD build. - Check the package using
R CMD check. Modify the package (if necessary) until no moreERRORSorWARNINGSoccur.
- Create a skeleton of the package using