9 R Interfaces
“You should try things; R won’t break.”
- Duncan Murdoch, from R-help (May 2016)
9.1 Introduction
R can be interfaced with non-native software packages or languages using a software binding procedure called an application programming interface (API)98. The binding provides glue code that allows R to work directly with foreign systems that extend its capacities. This can be done in two basic ways.
First, R-bindings for external, self-contained software programs can be used. This allows R-users to: 1) parameterize and initiate an external program using wrapper functions, and, 2) access the output from that program for further analysis and distillation. If one is using existing APIs, then these operations will generally not require knowledge of non-R languages (as the heavy lifting is being done with utility functions within particular R packages). One will, however, have to install the R package containing the API(s), and the software that one wishes to interface.
Second, one can harness useful characteristics of non-R languages by: 1) writing or utilizing source code for procedures in those languages, and 2) using APIs to run those processes in R, possibly following their compilation into entities called executable files (Section 9.1.6).
Although bindings for external software are considered briefly (Section 9.1.1), this chapter focuses primarily on interfaces of the second type, particularly bindings to the programming languages Fortran, C, C++, SQL, and Python. Brief backgrounds to those languages are provided here. These, however, should not be considered thorough introductions, given that: 1) I am not a computer language polyglot, and 2) my focus is to demonstrate how other languages can be interfaced with R, and not the languages themselves. Appropriate references to language resources are provided throughout the chapter99.
To account for the frequent use of distinct computer languages in this chapter, the following coloring conventions for chunk inputs will be used hereafter in this book100:
# R code9.1.1 R Bindings for Existing External Software
Many applications exist for interfacing R with extant, biologically-relevant software. For example, the R package arcgisbinding101 allows R-based geoprocessing within ArcGIS Pro and ArcGIS Enterprise.
Example 9.1 \(\text{}\)
Here I establish a connection to the ArcGIS software package on my computer from within R.
product: ArcGIS Pro (13.5.0.57366)
license: Advanced
version: 1.0.1.311 \(\blacksquare\)
The R package igraph (Csárdi et al. 2025) provides C-bindings for an extensive collection of graph-theoretic tools that can be applied in biological settings, e.g., K. Aho, Derryberry, et al. (2023). Wrappers for open-source bioinformatics software include the R package RCytoscape, from the Bioconductor repository, which allows cross-communication between the popular Java-driven software for molecular networks Cytoscape; the R package dartR.popgen which interfaces with C-based STRUCTURE software for investigating population genetic structure; and the R package strataG (currently only available on GitHub) which can interface with STRUCTURE, along with the bioinformatics apps: CLUMPP, MAFFT, GENEPOP, fastsimcoal, and PHASE.
R can also be accessed from popular commercial software. This capacity is particularly evident in commercial statistical software, including SAS, SPSS, and MINITAB.
9.1.2 Interfacing With Non-R Languages
Source code from other languages can often be interfaced to R at the command line prompt, and within R functions. For instance, we have already considered the use of Perl regex calls for managing character strings in Ch 4 (Section 4.3), and the R Markdown document processing workflow is largely a chain of Markup language conversions (Section 2.10.2.1). Other examples include code interfaces from C, Fortran, C++, SQL, and Python (all formally considered in this chapter), MATLAB (via package R.matlab, Bengtsson (2022)), and Java (via package rJava, Urbanek (2021))102.
9.1.2.1 Costs/Benefits of Interfacing Non-R Scripts
There are costs and benefits to creating/using interface scripts. Costs include:
- Scripts written in non-interpreted languages (e.g., C, Fortran, C++, see Section 9.1.6) will require compilation. Therefore it may be wise to limit such code to package-development applications (Ch 10) because R built-in procedures can facilitate this process during package building.
- Interfacing with older, low level languages (e.g., Fortran and C (Section 9.4)) increases the possibility for programming errors, often with serious consequences, including memory faults. That is, bugs bite (Chambers 2008)!
- Interfacing with some languages may increase the possibility for programs being limited to specific platforms.
- R programs can often be written more succinctly. For instance, Morandat et al. (2012) found that R programs are about 40% smaller than analogous programs written in C.
Despite these issues, there are a number of strong potential benefits. These include:
- A huge number of useful, well-tested applications have been written in other languages, and it is often straightforward to interface those procedures with R.
- The system speed of other languages may be much better than R for many tasks. For instance, looping algorithms written in non-interpreted languages, are often much faster than corresponding procedures written in R.
- Non-OOP languages may be more efficient than R with respect to memory usage.
9.1.3 Using RStudio as an IDE for other languages
RStudio can serve as a scripting IDE for many languages other other than R. These currently include: C, C++, CSS, JavaScript, Python, SQL, and Stan. To enable a particular IDE, one would go to File > New File in RStudio, and choose the desired format.
9.1.4 Interfacing with R using knitr Chunks
Language and program interfacing with R can be greatly facilitated with code chunks inserted in R Markdown or Sweave documents. This is because many languages other than R are supported by knitr and Sweave. Recall (Section 2.10.2.1.2) that the language engine for a particular knitr chunk is given by the first argument in the chunk options. For instance, in an R Markdown .rnw document ```{r } ``` initiates a conventional R code chunk, whereas ```{python }``` would begin a Python code chunk. In a Sweave .rnw document, one would specify a Python chunk using the header: <<engine = 'python'>>=. Here are the current knitr language engines.
names(knitr::knit_engines$get()) [1] "awk" "bash" "coffee" "gawk" "groovy"
[6] "haskell" "lein" "mysql" "node" "octave"
[11] "perl" "php" "psql" "Rscript" "ruby"
[16] "sas" "scala" "sed" "sh" "stata"
[21] "zsh" "asis" "asy" "block" "block2"
[26] "bslib" "c" "cat" "cc" "comment"
[31] "css" "ditaa" "dot" "embed" "eviews"
[36] "exec" "fortran" "fortran95" "go" "highlight"
[41] "js" "julia" "python" "R" "Rcpp"
[46] "sass" "scss" "sql" "stan" "targets"
[51] "tikz" "verbatim" "theorem" "lemma" "corollary"
[56] "proposition" "conjecture" "definition" "example" "exercise"
[61] "hypothesis" "proof" "remark" "solution" "glue"
[66] "glue_sql" "gluesql" Items 53-67 are not explicit computer languages, but allow specification of particular R Markdown document blocks, with appropriate labels and numbering. Note that knitr (and Sweave) engines extend to compiled languages including Fortran (engine = fortran), C (engine = c), see Section 9.4.0.1, and C++, via the Rcpp package (engine = Rcpp), see Section 9.5.1.
9.1.5 Source and Machine Code
Source code refers to human-readable instructions under the framework of some programming language. For instance, the script
[1] 3.3333is an example of R source code, with its evaluation result shown.
A computer, however, only fundamentally understands machine code (also called object code)103. Conventionally, machine code is a binary {0, 1} representation of a source code procedure (Section 12.2). The machine code for the R script x <- c(1, 3, 7); mean(x) is not show here. However, the binary (see Ch 12) translation of \(3.33\bar{3}\), is:
[1] 11.0101010101Source code must be translated into machine code before a computer can execute it.
9.1.6 Interpreted and Compiled Languages
R, along with many other useful languages (e.g., Python, JavaScript), is considered an interpreted language. In programming, an interpreter directly executes source code without the requirement of compilation. R uses a Scheme-like interpreter to translate source code into an intermediate representation of source code and machine code entities, which is then immediately executed. These operations generally rely on processes written in the language C. Because translation must precede machine code implementation, interpreted procedures tend to be slower than fully compiled procedures. This is particularly true for iterative processes like loops.
Non-interpreted (compiled) languages (for instance, Fortran, C, C++, C#, and Java) use a compiler (a conversion program) to transform some process, represented by source code, into machine code104. The compiler may also link to required external code and optimize program performance and cross-platform portability. The result of compilation is often called an executable file, or simply an executable (Figure 9.1). Executables can be called from within R (or elsewhere) to run independently, or to enhance other functions and procedures.
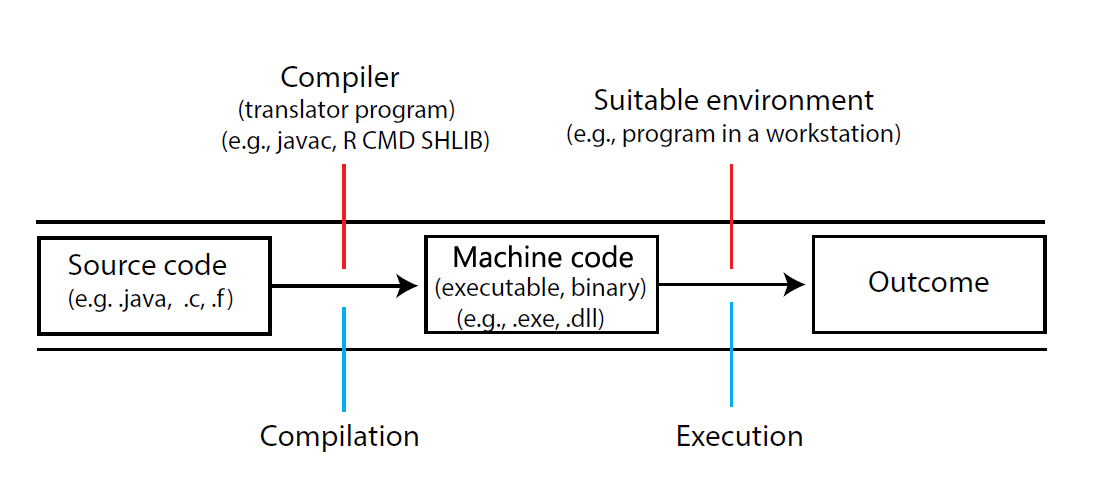
FIGURE 9.1: Creating an executable file in a compiled language.
Compilers are generally specific to underlying source code languages. For instance, the ILCPU compiler is intended for C# code, and the clisp compiler is intended for Lisp. The following compilation tools are very important to R-users. The last two are particularly important for Windows OS users.
-
The GNU Compiler Collection (GCC) contains a large number of open source compilers, including
gcc(for C),g++(for C++), andgfortran(for Fortran).
- MinGW (“Minimalist GNU for Windows”) is a free open source development environment for creating Windows applications. It includes a GCC port, along with other compilation tools specifically for Windows.
- Rtools is a Windows toolchain, intended primarily for building packages (and R) from source code. As of version 4.5, Rtools includes Msys2 –a collection of tools and libraries for building, installing and running Windows software, the GCC 14/MinGW-w64 compiler toolchain for Windows, and QPDF –a command line tool and C++ library that performs content-preserving transformations on PDF files.
9.2 Shells
Compiling object code will require installation/access to an appropriate compiler program. A compiler, in turn, will likely require initiation from a shell command line. Of course, Shells can be used for many purposes other than source code compilation105.
The Windows OS currently has two built-in command line shells106. The Command shell (also know as cmd.exe or cmd), was introduced in 1993 and maintains strong similarities to the venerable MS-DOS language framework (see Wikipedia (2025l)). PowerShell, introduced in 2006, is back-compatible with most cmd commands, but also has advanced programming features, including the ability to generate objects and handle OOP scripts. Other differences between cmd and PowerShell are discussed here. Processes for Windows shells differ in many respects from the POSIX (Portable Operating System Interface) compliant shells generally used by Unix-like systems. The most widely used POSIX shell, BASH107, allows straightforward execution and modification of Linux/Unix operations that may be difficult to translate to Windows OS108. The Windows Subsystem for Linux (WSL) allows one to run Linux, including BASH shells, directly on a Windows machine. I strongly recommend WSL for advanced shell procedures, including the generation of shell programs (Section 9.2.8 below), compilation of scripts using makefiles (Section 11.6), Windows management of R-driven apps in servers (Sections 11.5.7, 12.9.2), and high performance computing projects using R (Section 12.10.1).
Shell commands can be run directly from customized knitr chunks (see Section 9.1.4). For instance, BASH can be initiated (if available) using ```{bash }```, and the simpler Bourne POSIX shell can be initiated using ```{sh }```. In Windows, the R function shell() can be used to run shells, including cmd, sh, BASH, and others, directly from the R command line.
Most shells (including PowerShell, cmd, and BASH) allow auto-completion of commands using the Tab key. This is particularly convenient for completion of long file names. Further, like R-term, these shells allow scrolling through earlier commands using the \(\uparrow\) and \(\downarrow\) keys. To interrupt a system shell process, one can generally use Ctrl + C .
CAUTION!
Shells are powerful tools, and serious damage can be done to your computer through their misapplication. This is particularity true when running destructive commands, e.g., del, rmdir, format as an Administrator.
9.2.1 Simple Shell Procedures
Table 9.1 lists some simple shell commands, including a number that work the same way in both Windows and Linux/BASH.
| Operation | Windows | Linux/BASH |
|---|---|---|
| Change directory |
cd <path> go to <path>cd .. “up” one directorycd ~ (PS) or cd %HOMEPATH% (cmd) home directory cd \ root directory |
cd <path> go to <path>cd .. “up” one directorycd - previous directorycd ~ home directorycd / root directory |
| Root directive |
sudo (PS) |
sudo-s run specified shell |
| Print working directory |
pwd (PS) or cd (cmd) |
pwd |
| Clear shell | cls |
clear |
| List files in directory |
dir/B use bare format/O:D sort by timestamp /O:S sort by file size /O:E sort by file extension |
ls-t sort by timestamp-S sort by file size-X sort by file extension |
| Show directory nesting | trre |
tree |
Print <file>
|
type <file> (cmd) or cat <file> (PS) |
cat <file>-n number all output lines |
| Display text | echo |
echo |
Copy <file>
|
copy <file> <destination> |
cp <file> <destination> |
Move <file>
|
move <file> <destination> |
mv <file> <destination> |
| Print system details |
systeminfo/s IP address |
uname-a all details-p processor type-o operating system |
Find <string>
|
findstr <string>/I ignore case/V print non-matching lines |
grep <string> <path> -i ignore case -v print non-matching lines -c print only output line counts -l print only filename matches -E use extended regex -P use Perl-compatible regex-F interpret pattern as “fixed” (not regex) |
| Sort and print |
sort/unique return only unique outcomes |
sort-u return only unique outcomes |
| Help | help <command> |
help <command> or <command> --help
|
| Exit shell | exit |
exit |
Some additional details may be useful for comprehending Table 9.1. The root directory of a computer (obtained using cd \ in PowerShell and cmd, and cd / in BASH) is the hierarchical starting point for files. That is, it is the root of a computer’s directory tree. The root directory is managed by the so-called “superuser”, and can be used to implement system-wide changes. Commands at the root-level will often require sudo (superuser do) privileges, initiated with a password109. The home directory (obtained using cd ~ or cd %HOMEPATH% (cmd)) is located within the root directory, and comprises a personal directory system starting point110. Users will generally have full read/write access within their home directory, although a non-root sudo password may be required for some operations. When switching between BASH and Windows shells, it is important to remember that while Windows nests directories using backslashes, for instance Dir1\Dir2\Dir3, Unix-alike shells (and systems developed under Unix, like R) use forward slashes: Dir1/Dir2/Dir3.
User-specified options available for cmd procedures will generally be preceded with a forward slash, /, whereas user-specified options in BASH will be preceded with a dash,- (Table 9.1). For instance, the BASH ls procedure has an option -1 that causes one file to be printed per line, and an option -S that sorts by file and/or directory size. To run ls with those options one could use: ls -1S.
Note that methods for obtaining help differ by shell. To get help for a cmd or PowerShell process named <command> one could use: help <command>. BASH help can be obtained two ways. For a BASH built-in process named <command> one would use: help <command>. BASH internal commands include cd and pwd. However, for an external command or executable (e.g., grep, sort) one would use <command> --help. A formatted BASH manual for external processes can often be obtained using the format: man <command>.
The use of several metacharacters is consistent across shell languages (Table 9.2). Note that the command .\script can be used in PowerShell and BASH to run a script or program within the current working directory. This allows one to run executables that are not on defined Environmental Paths. If a program does exist on a path, then one can generally run it from a shell by simply typing program or, if appropriate, program.exe.
| Operation | Windows | Linux/Bash |
|---|---|---|
| Expand to match | * |
* |
| Redirect output | > |
> |
| Append output | >> |
>> |
| Redirect input | < |
< |
Call variable name
|
\%name\% (cmd) or \$name (PS) |
\$name |
| Pipe | \(\mid\) | \(\mid\) |
| Escape |
^ (cmd), \ (PS) |
\ |
Run script in current dir |
script (cmd), ./script (PS) |
./script |
Additional guidance for Windows shells can be found at the learn.microsoft.com website. Additional guidance for BASH can be found here.
Example 9.2 \(\text{}\)
To navigate to the root directory of my computer, I could type cd \ in Windows cmd or PowerShell, or type cd / in BASH (Table 9.1). Here I use cmd:
C:\>Note that the cmd shell has the same command line prompt as R: >. Now, however, the current directory is also included as part of the shell command line. This format (also used by PowerShell and BASH) occurs because the current directory will be a shell’s working directory.
\(\blacksquare\)
Example 9.3 \(\text{}\)
Here I access the Ubuntu version of Linux implemented in WSL from a Windows shell:
Welcome to Ubuntu 24.04.3 LTS (GNU/Linux 6.6.87.2-microsoft-standard-WSL2 x86_64)
/mnt/c/Users/ahoken$The BASH command line operator is $. The hierarchy: /mnt/c/Users/ is a conventional path for Linux in WSL. In particular, /mnt is a Linux location designation for “mounting” to other file systems or devices. The directory /c is the WSL mount point for the Windows C: drive.
Here I navigate to the Linux root directory:
/$To obtain shell superuser directory privileges, I can use:
/mnt/c/Users/ahoken#Because I have root access, the prompt is now #.
\(\blacksquare\)
Example 9.4 \(\text{}\)
To navigate to my home directory, I could use cd %HOMEPATH% in cmd, or use cd ~ in BASH or PowerShell. Here is result in cmd:
C:\Users\ahoken>Here I use PowerShell:
PS C:\Users\ahoken>Here I use BASH:
~$Here I print my (current) working directory in BASH:
/home/ahokenNote that this is not the same location as /mnt/c/Users/ahoken.
\(\blacksquare\)
Directories in shells can be accessed using absolute paths or shortcut relative paths. One can always navigate to a particular location by specifying its entire absolute path, starting at the root. For instance, if the full path to my desired location from the BASH root is: /mnt/c/Users, I could navigate to it using the absolute path: cd /mnt/c/Users. On the other hand, if I was already in the directory c, I could navigate to Users using the relative path: cd Users. If Dir1 was at the top-level within my home directory, I could navigate to it in BASH using cd ~/Dir1.
9.2.2 Plain Text Searches and Management
It is often straightforward to examine and manage files, particularly plain text files, from a shell. This can be facilitated through the use of shell wildcards. The most useful of these is the asterisk, *, which means: “expand to match the names of all files and directories in the current directory” (Table 9.2). Thus, in a shell environment, * is similar to the regex command .*.
Example 9.5 \(\text{}\)
What if I wanted to list all the R-Markdown files (those with an .rmd file extension) in the home directory for this book? I could navigate to the correct directory in cmd, and type dir /b *.rmd:
C:\> cd "C:\Users\ahoken\Documents\GitHub\Amalgam"
C:\Users\ahoken\Documents\GitHub\Amalgam> dir /b *.Rmd01-Ch1.Rmd
02-Ch2.Rmd
03-Ch3.Rmd
04-Ch4.Rmd
05-Ch5.Rmd
06-Ch6.Rmd
07-Ch7.Rmd
08-Ch8.Rmd
09-Ch9.Rmd
10-Ch10.Rmd
11-Ch11.Rmd
12-Ch12.Rmd
13-Appendix.Rmd
14-references.Rmd
Amalgam-of-R.Rmd
index.RmdRecall (Table 9.1) that dir lists working directory components in Windows OS (PowerShell can use either dir, or ls (like BASH)). The /b option in dir means: “use a bare format (no heading information or summary).” The metacharacter * serves as a wildcard (Table 9.2). Specifically, *.rmd indicates that only files in the working directory with a .rmd extension (whose names end with .rmd) should be listed. I could get the same result in BASH using:
\(\blacksquare\)
The * shell wildcard can be used in more sophisticated ways. For instance, to list all text files within all directories starting with D, I could use: ls D*\*.txt in PowerShell or ls D*/*.txt in BASH.
Example 9.6 \(\text{}\)
To search for the text: "An Amalgam of R" in the R Markdown document index.rmd, I could use the Windows shell findstr command:
title: "An Amalgam of R"Note that I escape both quotes and spaces in the string. The entire line of text containing the string is: title: "An Amalgam of R" and is part of the YAML header in the file index.rmd (Section 2.10.2.1). For more information type help findstr in cmd (Table 9.1).
\(\blacksquare\)
It may often be easier to wrangle text strings using regular expressions under BASH instead of base R (Section 4.3.3). This is because the need for sequential escape backslashes, e.g., \\ will be unnecessary unless one is actually escaping a backslash (see Section 4.3.3.4). Unfortunately, PowerShell, and particularly cmd, have limited regex handling capabilities.
Example 9.7 \(\text{}\)
Here I navigate to the home directory of this book using BASH:
Here I get an approximate count of the number of R Markdown R code chunks in the book by querying the string '{r' within underlying .Rmd files, using the grep option -c.
01-Ch1.Rmd:18
02-Ch2.Rmd:191
03-Ch3.Rmd:260
04-Ch4.Rmd:115
05-Ch5.Rmd:57
06-Ch6.Rmd:158
07-Ch7.Rmd:121
08-Ch8.Rmd:177
09-Ch9.Rmd:173
10-Ch10.Rmd:24
11-Ch11.Rmd:171
12-Ch12.Rmd:57
13-Appendix.Rmd:2
14-references.Rmd:0
Amalgam-of-R.Rmd:1529
index.Rmd:5There are over 1400 R chunks in the book (including some, hidden, formatting scripts)111, and 259 occur in Chapter 3. Here are the number of C++, SQL and Python chunks used in the current chapter:
342647\(\blacksquare\)
9.2.3 Redirect and Append
In cmd, PowerShell, and BASH, the > operator redirects output from a command to a user defined file (Table 9.2). The process will overwrite existing content with that file name. The >> operator appends output from a command to an existing file (Table 9.2). If the requested file does not exist, then >> is equivalent to >.
Example 9.8 \(\text{}\)
The BASH procedure below writes a result from the previous example (Example 9.7) to a new file called R_chunks.txt.
The first line in script below creates a new file called greeting.txt, which contains the text Hello, World!. The second line appends the additional text Goodbye, World! to the file.
\(\blacksquare\)
9.2.4 Creating and Calling Variables
BASH and cmd do not recognize objects with associated methods –although PowerShell is actually an an OOP scripting language. Nonetheless, all three approaches can be used to create and call variables.
- To create a variable in cmd named
NAME, with bound valueVALUE, I would use:set NAME=VALUE(no space before or after=). To call the variableNAME, I could then specify:%NAME%(Table 9.2). - To create a variable in PowerShell or BASH named
NAME, with bound valueVALUE, I would simply use:NAME=VALUE(no space before or after=). To callNAME, I could then use:$NAME(Table 9.2). Note thatNAMEwould be a local variable, only accessible in the current shell session or script.
Example 9.9 \(\text{}\)
Here is a simple cmd example:
Hello_World!There are many many built-in cmd variables, including as %USERNAME%, %PATH%, and %OS%.
My operating system is Windows_NT\(\blacksquare\)
Example 9.10 \(\text{}\)
In BASH or PowerShell, I could do something like:
Hello, World!Like cmd, BASH has many built-in variables. These include $HOME, $USER, and $HOSTTYPE. The latter prints a computer’s CPU type (see Section 12.1.1).
x86_64\(\blacksquare\)
9.2.5 The AWK and sed Unix Procedures
Aside from the grep utility, BASH has command line access to number of other useful Unix processes and programs. These include
-
AWK: A Unix scripting language named for its authors A. V. Aho, Kernighan, and Weinberger (2023). AWK is typically used as a text processing and data extraction and reporting tool. We would call AWK from BASH using the command
awk. Importantawkoptions include:-
-F'fs'usefsas a field separator. -
-iload an AWK source library.
-
-
sed (stream editor): A Unix utility for parsing a transforming text, and supporting regular expressions, that uses its own simple scripting language. We would call sed from BASH using the command
sed. Importantsedoptions include:-
-Eor-ruse extended regex. -
-fadd the contents of script-file to command. -
-eallow multiple sed commands.
-
AWK programs have optional designated BEGIN{} and END{} sections, and a required main section, also delimited by curly braces: {}.
Example 9.11 \(\text{}\)
Assume we have the following lines of comma separated text stored in the current directory as the file a_acid.txt. A copy of the file can be found at: https://amalgamofr.org/a_acid.txt
Ala
Arg
Asn
Asp
CysThe awk option -F is used to specify field (column) separators, -F',' indicates that commas separate columns. The simple script above does not contain optional BEGIN{} and END{} sections. The main section '{print $2}' indicates that the second column should be printed.
\(\blacksquare\)
Sed programs have the general structure: sed options 'command' input_file. Important sed commands include s, which precedes specific substitution instructions to be applied to the input_file.
Example 9.12 \(\text{}\)
The snippet s, below indicates that a substitution should be applied to a_acid.txt, which is a comma delimited file. The snippet , ,_, indicates that spaces in discrete strings should be replaced with the underscore character.
Alanine,Ala,A
Arginine,Arg,R
Asparagine,Asn,N
Aspartic_acid,Asp,D
Cysteine,Cys,C\(\blacksquare\)
9.2.6 Pipes
PowerShell, cmd, and BASH all allow invocation of the Unix-style pipe operator | (Table 9.2). Reflecting usage of the native R pipe |> (Section 5.2), the shell operation a | b means: “take the result from a and evaluate it in b.”
Example 9.13 \(\text{}\)
Here I continue Example 9.7 by counting the number of Example sections in the book by counting occurrences of the string '::: {#exm', using a pipe. To allow proper formatting, this string precedes every Example in each chapter .Rmd file.
620AWK is often used in Unix pipes. The awk option -F specifies that colons, :, should be viewed as column delimiters in the pipe stream. The command {n+=$2} means: “calculate and print the sum of values in the second column of the stream.” The command END {print n} ends the stream, and prints the cumulative sum stored in n (some object-oriented applications are possible in AWK).
\(\blacksquare\)
Example 9.14 \(\text{}\)
The open source software application mothur (Schloss et al. 2009) is often used for bioinformatics data processing. A mothur taxonomy file, which provides a hierarchical taxonomic classification of organisms in samples (e.g., domain, kingdom, phylum, class, order, family, genus, species), can have a very complex plain text format. Below are five lines of output from a mothur taxonomy file, from a recent project that investigated the microbiomes of intermediate streams in the intermountain west of the United States. Importantly, note that 1) taxonomic categories, designated d__, k__, etc. (and their assignments) are separated with semicolons, and 2) the number of assigned categories varies with taxon. For example, the taxa on lines 1, 2, 4, and 5 have genus (i.e., g__) as their finest level of taxonomic resolution. However, the taxon on line 3 has a species (__s) designation, albeit limited to uncultured_bacterium.
d__Bacteria; p__Bacteroidota; c__Bacteroidia; o__Cytophagales; f__Cyclobacteriaceae; g__uncultured
d__Bacteria; p__Planctomycetota; c__Planctomycetes; o__Gemmatales; f__Gemmataceae; g__uncultured
d__Bacteria; p__Myxococcota; c__bacteriap25; o__bacteriap25; f__bacteriap25; g__bacteriap25; s__uncultured_bacterium
d__Bacteria; p__Bacteroidota; c__Bacteroidia; o__Sphingobacteriales; f__Sphingobacteriaceae; g__Mucilaginibacter
d__Bacteria; p__Acidobacteriota; c__Acidobacteriae; o__Solibacterales; f__Solibacteraceae; g__Candidatus_SolibacterAssume that I have a plain text file, taxon.txt, containing the data above. And that the file is located in my home directory: PS C:\Users\ahoken>. A copy of the file exists at https://amalgamofr.org/taxon.txt.
To facilitate text analysis of these data I will use BASH. I navigate to my home directory using cd ~, and print the working directory using pwd to ensure that I am in the correct place.
/home/ahokenI verify that taxon.txt is present by listing files names in the directory that start with the string taxon by placing the * wildcard after taxon.
taxon.txt
taxon1.txt
taxon2.txtHere I list all phylum names (those that start with p__ and end with ;):
p__Bacteroidota;
p__Planctomycetota;
p__Myxococcota;
p__Bacteroidota;
p__Acidobacteriota; Recall (Section 4.3.3.5) that the Perl-compatible regular expression (PCRE) wildcard \b (\\b in R) connotes a word boundary, and that \w (\\w in R) represents a “word” character. The * indicates: “occurring 0 or more times”. The grep options -P and -o indicate: “use Perl regular expressions”, and “show only nonempty parts of lines that match”, respectively.
It might be nice to: 1) print phylum names without the p__ head and the semicolon tail, and 2) generate a list of phyla in the sample by dropping redundant phylum names (there are two Bacteroidota taxa).
Acidobacteriota
Bacteroidota
Myxococcota
PlanctomycetotaThe regular expression above has three components: a so-called positive lookbehind: '(?<=...)', a character class: '[...]', and a subsequent pipe process: |....
- The positive lookbehind,
'(?<=p__)', is a PCRE procedure. It requires the text immediately preceding the string of interest to bep__. The textp__, however, will not be included in the output stream. - Recall that a regex character class (Section 4.3.3.4) will be a text pattern, situated between square braces. Recall also (Section 4.3.3) that
^, when it occurs within braces, is the regex negation (not) operator. When used outside of braces^means: “start of string”. The (negated) character class'[^; ]', matches characters that are not semicolons (;) or spaces (). The+at the end of the class designation specifies: “one or more” occurrences of the defined pattern. - The pipe process,
| sort -u, sorts the preceding PRCE matched output and returns only unique strings (option-u).
\(\blacksquare\)
9.2.7 Customizing Shell Output
The BASH command head, provides the beginning components of requested output (similar to the R function head()). Its important options include:
+ -n NUM' print the number of lines given in NUM.
+ -v always print headers giving file names.
The head command is generally used in the context of a pipe.
Example 9.15 \(\text{}\)
The script below would display the first three files/directories in the current working directory.
01-Ch1.Rmd
02-Ch2.Rmd
03-Ch3.Rmd\(\blacksquare\)
In cmd, PowerShell, and BASH, the echo command can be used to improve/clarify shell output.
Example 9.16 \(\text{}\)
For example,
Here are the first three files in the currect directory:
01-Ch1.Rmd
02-Ch2.Rmd
03-Ch3.RmdOne can embed BASH processes (and their output) with echo text by writing those processes between the delimiters $( and ).
The first (alphanumeric) file in /c/Users/ahoken/Documents/GitHub/Amalgam is: 01-Ch1.Rmd\(\blacksquare\)
9.2.8 Shell Programs
Shells process and execute user-defined inputs one line at a time. One can, however, call shell batch scripts or programs, consisting of multiple commands. In BASH and PowerShell one would initiate this process with the characters: #!, followed by a path to the executable that would interpret the script. The entity #! is called a shebang. The name corresponds to sounds of its components: the “sh” sound from the word “hash mark”, and the “bang” from the exclamation point.
To enable and control the executability of Linux/Unix shell programs, the powerful utility chmod, which means (change file mode). The chmod command a indicates that all users will have access to a program. The chmod commands r,w, and x indicate that users can read, write, and execute a program respectively. Importantly, program changes invoked by chmod are permanent. That is, once the file mode is established, one does not need to re-invoke chmod for a program, unless the program or its permissions are changed.
Example 9.17 \(\text{}\)
Here is a script, to be interpreted in BASH, that will print the number of unique Phyla, Classes, Orders and Families, in an arbitrary text file with mothur taxonomy formatting.
#!/bin/bash
FILE_NAME="$1"
echo "Phyla = $(grep -Po '(?<=p__)[^; ]+' $FILE_NAME | sort -u | wc -l) "
echo "Classes = $(grep -Po '(?<=c__)[^; ]+' $FILE_NAME | sort -u | wc -l) "
echo "Orders = $(grep -Po '(?<=o__)[^; ]+' $FILE_NAME | sort -u | wc -l) "
echo "Families = $(grep -Po '(?<=f__)[^; ]+' $FILE_NAME | sort -u | wc -l) "- On Line 1, I initiate the program with
#! /bin/bash, allowing the source code to be interpreted in the BASH language. - On Line 2, I create a variable called
FILE_NAMEwhose value (file path and name) will be set by the user when calling the function. The ciphers$1-$9can be used to set the first nine arguments in a shell program.
- On Lines 4-7 I insert
greplookbehind processes (Example 9.14) in conjunction withechotext (see Example 9.16). These results are then sifted through two consecutive pipes, usingsort -u, and a “new” BASH commandwc -l, which prints newline counts.
The most widely used extension for shell scripts is .sh. One can create a file with a particular file extension with the R function file.create(). For example, the code file.create("foo.sh") creates a file named foo.sh that I can open (from my working directory) in a text editor (e.g., Notepad) or IDE (e.g., RStudio) to build a shell script. From a shell command line one could also use a redirect to create the file. For instance: > foo.sh.
I create/place the file taxaprog.sh within my home directory. To eliminate the possibility of hidden Windows formatting, I use one of the built-in Ubuntu text editors, nano, to open and edit the file by typing:
After building (via copy and paste) the shell script in nano, I end the nano session by entering Ctrl + X (to exit the session), Y (to indicate I want to use the same file name), and Enter to return to the BASH prompt.
The script is poised to run. However to make it executable (by all) I need to specify:
For verification, I apply the program to the to the taxon.txt dataset from Example 9.14. Note the use of ./ (Table 9.2).
I get:
\(\blacksquare\)
9.3 Compilation for R Interfacing
Windows and Mac OS executables will generally have an .exe or .app extension, respectively, whereas extensions for Linux/Unix files are not formally required for a file be recognized and run as an executable. For distribution in R packages, however, executables must have a shared library format, with .dll, .dylib, and .so, extensions for Windows, Mac OS, and Linux/Unix operating systems, respectively112. Shared library objects are different from conventional executables in that they cannot be evaluated directly. In this case, R will be required as the executable entry point.
R has its own shared library compilers for Fortran, C, and several other languages via its SHLIB procedure113, which is accessed from the Rcmd executable. The Rcmd program is located in the R bin directory, following a conventional download from CRAN, along with several other important R executables, including R.exe and Rgui.exe. Rcmd procedures are typically invoked from a system shell (e.g., cmd, PowerShell, BASH) using the format: R CMD <procedure> <args>. Here <procedure> is currently one of INSTALL, REMOVE, SHLIB, BATCH, build, check, Rprof, Rdconfig, Rdiff, Rd2pdf, Stangle, Sweave, config, open, and texify. The entity <args> defines options and arguments specific to an Rcmd <procedure>114. For example, the shell script:
or
or
would prompt the building of a shared library object from the user-defined script foo, with option -c, which would automatically remove files created during compilation. The foo script itself could be comprised of Fortran, C, C++, or C-Obj source code.
There are actually several ways to compile shared libraries for use in R.
- First, as noted above, one could compile a shared library from some script,
foo, by runningR CMD SHLIB fooat a shell command line. The shared library could then be loaded and called, using an appropriate foreign function interface, e.g.,.Call()(see Section 9.4). I apply this approach from the Windows Command shell in Example 9.18. - Second, one could rely only on knitr language engines (see Section 2.7 in Xie, Dervieux, and Riederer (2020)). In particular, one could write a script for a compiled language within a chunk with an appropriate language engine. The chunk would be automatically compiled using
SHLIBwhen running the chunk. The resulting shared library could then be loaded and called in a subsequent R chunk, using an appropriate foreign function interface. See caveats, however, in Section 9.4.0.1. - Third, one could use a non-
R CMD SHLIBcompiler; for instance, Windows GCC tools in Rtools. Rtools compilers are used throughout Section 9.5. The package inline uses the GCC to allow users to create, compile, and run scripts, written in compiled languages, all from the R command line (see Sections 9.5.1 and 9.5.2).
9.4 Fortran and C
S, the progenitor of R, was created at a time when Fortran routines dominated numerical programming, and R arose when C was approaching its peak in popularity. As a result, strong connections to those languages, particularly C, remain in R115. R contains specific base functions for interfacing with both C and Fortran executables: .C() and .Fortran(). A more recently developed function, .Call(), which allows straightforward exchanges of SEXP objects to and from C, is formally introduced in Section 9.5.
Recall that an R object of class numeric will be automatically assigned to base type double, although it can be easily coerced to base type integer (with information loss through the elimination of its “decimal” component).
as.integer(2.5)[1] 2Many other languages, however, do not automatically assign base types. Instead, explicit user-assignments for underlying base types are required.
If one is interfacing R with Fortran or C, only a limited number of base types are possible (Table 9.3), and one will need to use appropriate coercion functions for R objects if one wishes to use those objects in Fortran or C scripts116, when using .C() and .Fortran(). Interfaced C script arguments must be pointers, and arguments in Fortran scripts must be arrays for the types given in Table 9.3.
| R base type | R coercion function | C type | Fortran type |
|---|---|---|---|
logical |
as.integer() |
int * |
integer |
integer |
as.integer() |
int * |
integer |
double |
as.double() |
double * |
double precision |
complex |
as.complex() |
Rcomplex * |
double complex |
character |
as.character() |
char ** |
character*255 |
raw |
as.character() |
char * |
none |
Raw Fortran source code is generally saved as an .f, or (.f90 or .f95; modern Fortran) file, whereas C source code is saved as an .c file. One could create the files foo.f90 and foo.c with file.create("foo.f90") and file.create("foo.c"), respectively. RStudio provides an IDE for C, allowing straightforward generation and editing of .c files.
9.4.0.1 Running C and Fortran code directly from chunks
As noted in Section 9.3, C and Fortran scripts can be run directly from customized knitr chunks (Section 9.1.4). Specifically, C source code can be specified using ```{c }``` in the chunk header, whereas Fortran source code can be specified using ```{fortran }``` or ```{fortran95 }``` (modern Fortran). The source code in chunks will be run through a compiler resulting in a shared library executable. For instance, the knitr engine c: 1) saves the chunk source code to a temporary .c file, 2) compiles the source code into a shared library using gcc (requiring installation of Rtools), and 3) loads the shared object as an .so file (Unix-alikes) or a .dll file (Windows).
The process above may be hampered by a number of factors117, including non-Administrator permissions and missing or incorrect Environmental Path definitions. As a result, I describe a more conventional process in Section 9.4.1 below, involving separate steps for C and Fortran source code creation, compilation, and execution.
9.4.1 Writing, Compiling and Executing C and Fortran Programs for R
Notably, native R SHLIB compilers will only work for Fortran code written as a subroutine118 and C code written in void formats119. As a result, neither code type will return a value directly.
Depending on Environmental Paths that have been established for R on your computer, different programmatic approaches may be necessary to implement R CMD SHLIB foo, where foo user-defined source code file.
- In the simplest scenario, one could use
R CMD SHLIB foofrom a system shell, after navigating to the system location offoo, or useR CMD SHLIB <path to foo>foofrom any location where<path to foo>is the absolute path tofoo. - If no Environmental Path has been set for
Rcmdprocesses, one could navigate to the R bin directory and useR CMD SHLIB <path to foo>foo(see Example 9.18 below) or<path to Rcmd>R CMD SHLIB <path to foo>foofrom any location where<path to Rcmd>is the absolute path to theRcmdprogram. - In the worst case scenario, one would have to physically copy or move
footo the R bin directory (generally requiring Administrator privileges) before applyingR CMD SHLIB foo.
Example 9.18 \(\text{}\)
Here is a simple example for calling Fortran and C compiled executables from R to speed up looping. The content follows class notes created by Charles Geyer at the University of Minnesota. Clearly, the example could also be run without looping. Equation (9.1) shows the simple formula for converting temperature measurements in degrees Fahrenheit to degrees Celsius.
\[\begin{equation} C = 5/9(F - 32) \tag{9.1} \end{equation}\]
where \(C\) and \(F\) denote temperatures in Celsius and Fahrenheit, respectively.
Here is a Fortan subroutine for calculating Celsius temperatures from a dataset of Fahrenheit measures, using a loop.
subroutine FtoC(n, x)
integer n
double precision x(n)
integer i
do 100 i = 1, n
x(i) = (x(i)-32)*(5./9.)
100 continue
endThe Fortran code above consists of the following steps:
- On Line 1 a subroutine is invoked using the Fortran function
subroutine. The subroutine is namedFtoC, and has argumentsx(the Fahrenheit temperatures) andn(the number of temperatures) - On Line 2 the entry given for
nis defined to be an integer (Table 9.3). - On Line 3 we define
xto be a double precision numeric vector of lengthn. - On Line 4 we define that the looping index to be used,
i, will be an integer. - On Lines 5-7 we proceed with a Fortran
doloop. The codedo 100 i = 1, nmeans that the loop will 1) run initially up to 100 times, 2) have a lower limit of 1, and 3) have an upper limit ofn. The code:x(i) = (x(i)-32)*(5./9.)calculates Eq. (9.1). The code5./9.is used because the result of the division can be a non-integer. The code100 continueallows the loop to continue ton. - On Line 8 the subroutine ends. All Fortran scripts must end with
end.
I save the code under the filename FtoC.f90, and transfer it to an appropriate directory (I use C:/Users/ahoken/Documents/Amalgam/Amalgam_Bookdown/scripts/). I then open a Windows shell editor.
I compile FtoC.f90 using the script R CMD SHLIB FtoC.f90. Thus, in the cmd shell (command line simplified for clarity) I enter:
> cd C:\Program Files\R\R-4.4.2\R\bin\x64
> R CMD SHLIB C:/Users/ahoken/Documents/GitHub/Amalgam/scripts/FtoC.f90Note the change from backslashes to (Unix-style) forward slashes when specifying addresses for SHLIB. The command above creates the compiled Fortran executable FtoC.dll. Specifically, the Fortran compiler, gfrotran, from within the GCC, is used to create an intermediate object file, FtoC.o. The object file is then used to create a .dll file with the gcc program. By default, the .dll is saved in the directory that contained the source code. Finalization of the compilation requires linkage to the the RTools MinGW toolchain.
Steps in the compilation process can be followed (with some difficulty) in the cmd shell output below. Some lines are broken to increase clarity.
using Fortran compiler: 'GNU Fortran (GCC) 14.2.0'
gfortran -O2 -mfpmath=sse -msse2 -mstackrealign
-c C:/Users/ahoken/Documents/GitHub/Amalgam/scripts/FtoC.f90
-o C:/Users/ahoken/Documents/GitHub/Amalgam/scripts/FtoC.o
gcc -shared -s -static-libgcc
-o C:/Users/ahoken/Documents/GitHub/Amalgam/scripts/FtoC.dll
tmp.def C:/Users/ahoken/Documents/GitHub/Amalgam/scripts/FtoC.o
-LC:/rtools45/x86_64-w64-mingw32.static.posix/lib/x64
-LC:/rtools45/x86_64-w64-mingw32.static.posix/lib -lgfortran -lquadmath
-LC:/PROGRA~1/R/R-45~1.1/bin/x64 -lR In the output above, snippets beginning with -, define gfortran and gcc program options from within the GCC. For instance, -c means “compile and assemble, but do not link,” -o <file> means “place output in a defined <file>”, and -L<directory> links <directory> to the program search path. The -O family of flags (including -O0, -O1, and -O2) concern compilation optimization. The option -O2 indicates “high optimization” at the cost of longer compilation times. Importantly, the option -shared indicates that a shared library should be assembled instead of a standard executable. Details on many gcc (and gfortran) options can obtained by calling gcc --help from the BASH command line. The options -mfpmath, -msse2, -mstackrealign are so-called “target-specific options.” Details concerning those options are provided in gcc --help=target.
Here is analogous C loop script for converting Fahrenheit to Celsius.
void ftocc(int *nin, double *x)
{
int n = nin[0];
int i;
for (i=0; i<n; i++)
x[i] = (x[i] - 32) * 5. / 9.;
}The C code above consists of the following steps.
- Line 1 is a line break.
- On Line 2 a
voidfunction is initialized with two arguments. The codeint *ninmeans “access the value thatninpoints to and define it as an integer.” The codedouble *xmeans: “access the value thatxpoints to and define it as double precision.” - Lines 8-9 define the C
forloop. These loops have the general format:for ( init; condition; increment ) {statement(s); }. Theinitstep is executed first and only once. Next theconditionis evaluated. If true, the loop is executed. The syntaxi++literally means:i = i + 1. Note that code lines are ended with a semicolon,:and that indices (e.g.,i) start at0. Consideration of the C language is greatly expanded in Section 9.5, which considers the language C++.
Once again, I save the source code, FtoCc.c, within an appropriate directory. I compile the code using the command R CMD SHLIB FtoCc.c. Thus, at the at the Windows Command shell I enter:
> cd C:\Program Files\R\R-4.5.1\bin\x64
> R CMD SHLIB C:/Users/ahoken/Documents/GitHub/Amalgam/scripts/FtoCc.cThis creates the shared library executable FtoCc.dll.
using C compiler: 'gcc.exe (GCC) 14.2.0'
gcc -I "C:/PROGRA~1/R/R-45~1.1/include"
-DNDEBUG -I "C:/rtools45/x86_64-w64-mingw32.static.posix/include"
-O2 -Wall -std=gnu2x -mfpmath=sse -msse2 -mstackrealign
-c C:/Users/ahoken/Documents/GitHub/Amalgam/scripts/FtoCc.c
-o C:/Users/ahoken/Documents/GitHub/Amalgam/scripts/FtoCc.o
gcc -shared -s -static-libgcc
-o C:/Users/ahoken/Documents/GitHub/Amalgam/scripts/FtoCc.dll
tmp.def C:/Users/ahoken/Documents/GitHub/Amalgam/scripts/FtoCc.o
-LC:/rtools45/x86_64-w64-mingw32.static.posix/lib/x64
-LC:/rtools45/x86_64-w64-mingw32.static.posix/lib
-LC:/PROGRA~1/R/R-45~1.1/bin/x64 -lRBelow is an R-wrapper that can call the Fortran executable, call = "Fortran", the C executable, call = "C", or use R looping, call = "R". Several new functions are used. On Line 10 the function dyn.load() is used to load the shared Fortran library file FtoC.dll, while on Lines 14-15 dyn.load() loads the shared C library file FtoCc.dll. Note that the variable nin is pointed toward n, and x is included as an argument in dyn.load() on Line 15. On Line 11 the function .Fortran() is used to execute FtoC.dll, and on Line 16 .C() is used to execute FtoCc.dll.
F2C <- function(x, call = "R"){
n <- length(x)
if(call == "R"){
out <- 1:n
for(i in 1:n){
out[i] <- (x[i] - 32) * (5/9)
}
}
if(call == "Fortran"){
dyn.load("C:/Users/ahoken/Documents/Amalgam/Amalgam_Bookdown/scripts/FtoC.dll")
out <- .Fortran("ftoc", n = as.integer(n), x = as.double(x))
}
if(call == "C"){
dyn.load("C:/Users/ahoken/Documents/Amalgam/Amalgam_Bookdown/scripts/FtoCc.dll",
nin = n, x)
out <- .C("ftocc", n = as.integer(n), x = as.double(x))
}
out
}Here I create \(10^8\) potential Fahrenheit temperatures that will be converted to Celsius using (unnecessary) looping.
[1] 72.849 16.560 87.114 68.725 81.543 75.094Note first that the Fortran, C, and R loops provide identical temperature transformations. Here are first 6 transformations:
head(F2C(x[1:10], "Fortran")$x)[1] 22.6940 -8.5779 30.6188 20.4027 27.5241 23.9411
head(F2C(x[1:10], "C")$x)[1] 22.6940 -8.5779 30.6188 20.4027 27.5241 23.9411
head(F2C(x[1:10], "R"))[1] 22.6940 -8.5779 30.6188 20.4027 27.5241 23.9411However, the run times are dramatically different120. The C executable is much faster than R, and the venerable Fortran executable is even faster than C!
system.time(F2C(x, "Fortran")) user system elapsed
0.69 0.08 0.80
system.time(F2C(x, "C")) user system elapsed
0.70 0.09 0.78
system.time(F2C(x, "R")) user system elapsed
5.83 0.27 6.28 \(\blacksquare\)
9.5 C++
C++ (pronounced see plus plus) is a high-level, general-purpose, programming language that is well known for its simplicity, efficiency, and flexibility121. C++ was originally intended to be a mere extension of C. Although its present scope greatly exceeds this goal, C++ syntax remains similar to C. For instance, like C:
- C++ is a compiled language (it requires a compiler to convert its source code to an executable).
- Lines of C++ code end with semicolons,
;. - C++ comment annotations begin with
\\. - The
forloop syntax for C++ is:for (init; condition; increment). - C++ index values start at
0, meaning that the last index value will ben - 1. - Square braces,
[], can be used for subsetting. Although, see content regarding Rcpp C++ types below. - C++ logical operators are similar to those used in R. For example,
==is the Boolean equals operator,!is the unary operator for not, and the operators for and and or are&&and||, respectively. - Like C, C++ Boolean designations,
trueandfalseare used (instead ofTRUEandFALSE).
The major difference between C and C++ is that C++ supports objects and object classes, whereas C does not. Helpful online C++ tutorials and references can be found at https://www.learncpp.com/ and https://en.cppreference.com/w/cpp, respectively. As advanced resources, Wickham (2019) recommends the books Effective C++ (Meyers 2005) and Effective STL (Meyers 2001)122.
9.5.1 Rcpp
The R package Rcpp (Eddelbuettel 2013; Eddelbuettel and Balamuta 2018; Eddelbuettel et al. 2023) provides an extension of the R API, with a consistent set of C++ classes (Eddelbuettel and François 2023). As a result, the package allows users to employ the many useful characteristics of C++ –including fast loops, efficient calls to functions, and access to advanced data container classes including maps123 and double-ended queues124, while enjoying the benefits of R –including terse scripting and straightforward manipulation of vectors and matrices. As Wickham (2019) notes:
“I do not recommend using C for writing new high-performance code. Instead write C++ with Rcpp. The Rcpp API protects you from many of the historical idiosyncracies of the R API, takes care of memory management for you, and provides many useful helper methods.”
Useful resources for Rcpp include extensive vignettes from the package itself, Chapter 25 from Wickham (2019), and the online document Rcpp for everyone (Tsuda 2020).
In order to use Rcpp, users will require additional toolchains, including a dedicated C++ compiler.
- Windows users will need Rtools. Use of Rtools will require that its installation be along an defined environmental path.
- Mac-OS users will require Xcode command line tools.
- Linux users can use
sudo apt-get install r-base-devin a system shell like BASH.
Here I verify the presence of the gcc compiler in Rtools on my machine:
Sys.which('gcc') gcc
"C:\\rtools45\\X86_64~1.POS\\bin\\gcc.exe" C++ source code can be directly compiled and implemented (under Rcpp) using customized knitr chunks (Section 9.1.4). This requires the specification: ```{Rcpp }``` in the chunk header.
Example 9.19 \(\text{}\)
As a first step, Eddelbuettel and Balamuta (2018) recommend running a minimal example to ensure that the Rcpp toolchain is working. For instance:
[1] 4Here the function Rcpp::evalCpp() creates a compiled C++ shared library, specified in evalCpp(), from the text string "2 + 2". This step is accomplished via the function Rcpp::cppFunction() (see Example 9.22 below). The evalCpp() function then calls the shared library, using .Call(), to obtain a result via R.
\(\blacksquare\)
9.5.1.1 Data Types
Recall (Section 2.3.7) that R base types correspond to a C typedef alias called an SEXP (S-expression). Rcpp provides dedicated C++ classes for most of the 24 SEXP types. Some of these are shown– for scalar, vector, and matrix frameworks– in Table 9.4. Scalars can be aptly handled with C++ standard library, std, procedures. The Rcpp::Vector types are similar to std::vector125, although the former are designed to facilitate interactivity with R.
| R type | C++ (scalar) | Rcpp (scalar) | Rcpp::Vector |
Rcpp::Matrix |
|---|---|---|---|---|
logical |
bool |
- |
LogicalVector |
LogicalMatrix |
integer |
int |
- |
IntegerVector |
IntegerMatrix |
numeric |
double |
- |
NumericVector |
NumericMatrix |
complex |
complex |
Rcomplex |
ComplexVector |
ComplexMatrix |
character |
char |
String |
CharacterVector |
CharacterMatrix |
Date |
- |
Date |
DateVector |
- |
POSIXct |
time_t |
Datetime |
DatetimeVector |
- |
Rcpp also has types for R base types list and S4, and R class dataframe. These are called using Rcpp::List, Rcpp::S4, and Rcpp::Dataframe, respectively. Rcpp types are designated with their class names.
Example 9.20 \(\text{}\)
The code (not run) below creates Rcpp::Vector objects called v. Corresponding R code is commented above C++ code.
// v <- rep(0, 3)
NumericVector v (3);
// v <- rep(1, 3)
NumericVector v (3,1);
// v <- c(1,2,3)
// [[Rcpp::plugins("cpp11")]]
NumericVector v = {1,2,3};
// v <- 1:3
IntegerVector v = {1,3};
// v <- as.logical(c(1,1,0,0))
LogicalVector v = {1,1,0,0};
// v <- c("a","b")
CharacterVector v = {"a","b"};Note that curly braces {} are used to initialize the NumericVector object on Line 9, and the IntegerVector, LogicalVector, and CharacterVector objects on Lines 12, 15, 18, respectively. This reflects C++ 11 grammar126. C++11 can be enabled with the comment: // [[Rcpp::plugins("cpp11")]] (Line 8).
Here I create Rcpp::Matrix objects named m:
// m <- matrix(0, nrow=2, ncol=2)
NumericMatrix m(2);
// m <- matrix(v, nrow=2, ncol=3)
NumericMatrix m( 2, 3, v.begin());The matrix object on Line 4 above is filled using a Vector object named v. This is facilitated with the Rcpp Vector member function begin() (Section 9.5.1.2).
Below is a Rcpp::Dataframe with columns comprised of Vectors named v1 and v2.
Here is a Rcpp::List containing Vectors v1 and v2.
\(\blacksquare\)
9.5.1.2 Member Functions
Rcpp has useful C++ member functions (functions that can be used to interact with data of specific user-defined types) for its Vector, Matrix, List and Dataframe types. Specifically, for a member function foo that corresponds to a type defined for an object bar, I would run foo on bar by typing bar.foo(). Note that Rcpp member functions in Table 9.5 with generic names, e.g., length() are analogous to R methods for particular S3 and S4 classes (Section 8.7).
| Function | Vector |
Matrix |
Dataframe |
List |
Operation |
|---|---|---|---|---|---|
length(), size()
|
X | X | X | Returns length of List or Vector, or number of Dataframe columns |
|
names() |
X | X | X | Names attribute | |
sort() |
X | X | Sorts object into ascending order | ||
get_NA() |
X | X | Returns NA values |
||
is_NA(x) |
X | X | Returns True if a Vector element specified by x is NA
|
||
nrows() |
X | X | Returns number of rows | ||
ncols() |
X | X | Returns number of columns | ||
begin() |
X | X | X | Returns iterator pointing to first element | |
end() |
X | X | X | Returns iterator pointing to end of object | |
fill_diag(x) |
X | Fill Matrix diagonal with scalar x
|
9.5.1.3 Math with R-like Functions
Rcpp contains R-like functions that extend C++ std mathematical procedures evaluated under the C <math.h> header file, or the C++ <cmath> header. The Rcpp functions allow users to capitalize on the vectorized efficiencies of R, within C++ scripts, while using R-like grammar. Table 9.6 shows simple mathematical operators and functions that are generally applicable to both scalar and Rcpp::Vector objects. Conversely, Table 9.7 shows vectorized R-like functions from Rcpp, without analogues in <math.h>.
| Operation | C++ scalar |
Rcpp Vector
|
Description |
|---|---|---|---|
| addition | s1 + s2 |
v + s or v1 + v2
|
scalar or vector (elementwise) addition |
| subtraction | s1 - s2 |
v - s or v1 - v2
|
scalar or vector (elementwise) division |
| multiplication | s1 * s2 |
v * s or v1 * v2
|
scalar or vector (elementwise) division |
| division | s1 / s2 |
v / s or v1 / v2
|
scalar or vector (elementwise) division |
| modulo | s1 % s2 |
remainder of division of s1 by s2
|
|
| \(\mid x \mid\) | abs(s) |
abs(v) |
absolute value(s) of s or elements in v. |
| round | round(s,d) |
round(v,d) |
rounds s or elements in v to d digits. |
| \(\sqrt{x}\) | sqrt(s) |
square root of s
|
|
| \(\log_2\) | log2(s) |
\(\log_2\) of s. |
|
| \(\log_e\) | log(s) |
log(v) |
\(\log_e\) of s or elements in v. |
| \(\log_{10}\) | log10(s) |
log10(v) |
\(\log_{10}\) of s or elements in v. |
| \(e^x\) | exp(s) |
exp(v) |
\(\exp()\) of s or elements in v. |
| \(x^n\) | pow(s, n) |
pow(v,n) |
raises s or elements in v to nth power. |
| \(\sin(x)\) | sin(s) |
sin(v) |
sine of s or elements in v. |
| \(\cos(x)\) | cos(s) |
cos(v) |
cosine of s or elements in v. |
| \(\tan(x)\) | tan(s) |
tan(v) |
tangent of s or elements in v. |
| \(\text{asin}(x)\) | asin(s) |
asin(v) |
arcsine of s or elements in v. |
| \(\text{acos}(x)\) | acos(s) |
acos(v) |
arccosine of s or elements in v. |
| \(\text{atan}(x)\) | atan(s) |
atan(v) |
arctangent of s or elements in v. |
\(\text{}\)
| Operation |
Rcpp Vector, v
|
Description |
|---|---|---|
| \(\min(x)\) | min(v) |
minimum value of v
|
| \(\max(x)\) | max(v) |
maximum value of v
|
| \(\sum_{i=1}^n x_i\) | sum(v) |
sum of v
|
| cumulative sum | cumsum(v) |
cumulative sum of v
|
| cumulative product | cumprod(v) |
cumulative product of v
|
| range | range(v) |
min and max of v
|
| \(\bar{x}\) | mean(v) |
mean of v
|
| \(\tilde{x}\) | median(v) |
median of v
|
| \(s\) | sd(v) |
standard deviation of v
|
| \(s^2\) | var(v) |
variance of v
|
| C++ version of R function | sapply(v,fun) |
applies C++ function fun to v
|
| C++ version of R function | lapply(v,fun) |
applies C++ function fun to v; returns List
|
| C++ version of R function | cbind(x1, x2,...) |
combines Vector or Matrix in x1, x2
|
| C++ version of R function | na_omit(v) |
returns Vector with NA elements in v deleted |
| C++ version of R function | is_na(v) |
labels NA elements in v TRUE
|
It is important to note that C++, like many other languages including C, and Fortran Python will often generate integer results from mathematical operations, even though they should be double precision. This can be readily demonstrated using Rcpp::evalCpp().
Example 9.21 \(\text{}\)
Clearly the answer to \(\frac{5}{2}\) is \(2.5\). However, running this operation in C++ produces:
evalCpp("5/2")[1] 2One way around this is to add a decimal to the end of the 5 and 2, to indicate that integer division should not be applied. Revisit Example 9.18 for Fortran and C examples of this approach.
evalCpp("5./2.")[1] 2.5\(\blacksquare\)
9.5.1.4 Inline C++ Code
The function Rcpp::cppFunction() allows users to specify C++ code for a single function as a character string at the R command line (see minimal Example 9.19 above). The function compiles C++ code, and creates a link to the resulting shared library. It then defines an R function that uses .Call() to invoke the shared library.
Example 9.22 \(\text{}\)
Here is a simple function for generating numbers from a Fibonacci sequence. See Question 6 in the Exercises from Ch 8.
cppFunction(
'int fibonacci(const int x) {
if (x == 0) return(0);
if (x == 1) return(1);
return (fibonacci(x - 1)) + fibonacci(x - 2);
}')- On Line 2, the C++ function name
finbonacciis defined. The function output and the class of the argumentxare both defined to beint(integers). - On Lines 3-4 the first two numbers in the sequence are defined based on Boolean operators.
- On Line 5, later numbers in the sequence (\(n > 2\)) are defined.
The result from the script is an R function that loads the compiled shared library, based on the C++ function fibonacci, using .Call().
fibonaccifunction (x)
.Call(<pointer: 0x00007ff9ab3d1860>, x)Here we use the R function to generate the 10th Fibonacci number.
fibonacci(10)[1] 55\(\blacksquare\)
The R function Rcpp::sourceCpp()allows general compilation of C++ scripts that may contain multiple functions.
9.5.1.5 Formal C++ Scripts
We can use Rcpp to facilitate the creation of more conventional C++ scripts (not just character strings of C++ code). These will have the general form (Tsuda 2020):
#include <Rcpp.h>
using namespace Rcpp;
// [[Rcpp::export]]
RETURN_TYPE FUNCTION_NAME(ARGUMENT_TYPE ARGUMENT){
//function contents
return RETURN VALUE;
}- On Line 1, the code
#include <Rcpp.h>loads the Rcpp header fileRcpp.h. In several C-alike languages (C, C++, C-obj), header files can be use to provide definitions for functions, variables, and (in the case of C++) new class definitions (Table 9.4). See Chapter 6 in R Core Team (2024c). - The (optional) code
using namespace Rcpp(Line 2) allows direct access to Rcpp classes and functions. Without this designation, an Rcpp function or classfoowould require the callRcpp::foo, instead of simply,foo. - The comment:
// [[Rcpp::export]]:(Line 4) serves as a compiler attribute, and demarks the beginning of C++ code that will be accessible from R. TheRcpp::exportattribute is required (by Rcpp) for any C++ script to be run from R. The attribute currently requires specification as a comment, because it will be unrecognized within most compilers. - For
RETURN_TYPE FUNCTION_NAME(ARGUMENT_TYPE ARGUMENT){(Line 5) users must specify data types of functions, a function name, argument types, and arguments. -
return RETURN VALUE;is required if function output is desired.
As before, this process compiles the C++ code into shared library, and creates an R function (with the same name as the C++ function) that calls the shared library (Example 9.22). In knitr chunks, one can check and debug this process by calling R to run this function from within the chunk containing C++ scripts, using the code format:
where FUNCTION_NAME is the name of the resultant R function.
Example 9.23 \(\text{}\)
RStudio provides an IDE for C++ scripts. Further, a C++ file obtained using File>New File>C++ contains an example Rcpp-formatted C++ example function. The function, named timesTwo, multiplies some number by two:
#include <Rcpp.h>
using namespace Rcpp;
// [[Rcpp::export]]
NumericVector timesTwo(NumericVector x) {
return x * 2;
}Note use of the Rcpp type NumericVector to define function output and values for the argument, x (Line 5).
Running the code above compiles timesTwo into a shared library, and creates an R function (with the same name) in the global environment. This function loads the shared library for use in R.
timesTwo(5)[1] 10\(\blacksquare\)
Example 9.24 \(\text{}\)
As a series of biological examples, we will create C++ functions (using Rcpp tools) for measuring the diversity of ecological communities. Below is a function for calculating relative abundances of species in a community (individual species abundance divided by the sum of species abundances).
#include <Rcpp.h>
using namespace Rcpp;
// [[Rcpp::export]]
NumericVector relAbund(NumericVector x) {
int n = x.length();
double total = 0;
for(int i = 0; i < n; ++i) {
total += x[i];
}
NumericVector rel = x/total;
return rel;
}The function relAbund is a mixture of standard C++ code and calls to C++ classes and procedures from Rcpp. In particular,
- On Lines 1 and 2, I bring in the
Rcpp.hheader file, and load the Rcpp namespace. - On Line 4, I include the comment,
// [[Rcpp::export]]to prompt R to recognize code below the line. - On Line 6, I specify the data types of the function output,
NumericVector, the function name, the data type for the argumentNumericVector, and the argument itself,x. - Lines 7-8 are preliminary steps for the loop codified on Lines 9-11. On Line 7, an integer object
nis defined as the number of observations inx. This is done with the RcppVectormember functionlength()(Table 9.5) with the callx.length(). - Lines 9-11 comprise a standard C/C++ looping approach for calculating total abundance (the sum of
x). The useful operator+=adds the right operand to the left operand and assigns the result to the left operand. - On Lines 12-13 relative abundances are calculated and the resulting
NumericVectoris returned.
Recall (Example 6.18) that the dataset vegan::varespec describes the abundance of vascular plants, mosses, and lichen species for sites in a Scandinavian taiga/tundra ecosystem. Here I run the function for the site represented in row 1 (site 18).
[1] 0.00616592 0.12477578 0.00000000 0.00000000 0.19955157 0.00078475
[7] 0.00000000 0.00000000 0.01793722 0.02320628 0.00000000 0.01816143
[13] 0.00000000 0.00000000 0.05235426 0.00022422 0.00145740 0.00000000
[19] 0.00145740 0.00134529 0.00000000 0.24360987 0.24069507 0.03923767
[25] 0.00336323 0.00201794 0.00257848 0.00280269 0.00280269 0.00257848
[31] 0.00000000 0.00000000 0.00089686 0.00022422 0.00022422 0.00000000
[37] 0.00134529 0.00022422 0.00695067 0.00022422 0.00000000 0.00000000
[43] 0.00280269 0.00000000I ensure that the C++ shared library relAbund views varespec[,1] as double precision by specifying mode = "double" in as.vector().
Recall (Example 8.22) that species relative abundances are used in calculating measures of \(\alpha\)-diversity. The code below calculates Simpson diversity (Eq. (8.4)) from a vector of abundance data.
#include <Rcpp.h>
#include <cmath>
using namespace Rcpp;
// [[Rcpp::export]]
double simpson(NumericVector x) {
NumericVector y = na_omit(x);
double total = sum(y);
NumericVector relsq = pow(y/total, 2);
return 1 - sum(relsq);
}Note that on Line 7, I have dramatically simplified the calculation of relative abundance by replacing the for loop in relAbund with the R-like Vector function Rcpp::sum() (Table 9.6). Other R-like C++ functions used above include na_omit() (Line 6) Rcpp::pow() and (Line 8). The former allows handling data with missing values.
simpson(as.vector(varespec[1,], mode = "double"))[1] 0.82171\(\blacksquare\)
Example 9.25 \(\text{}\)
The code below shows how one would run some simple mathematical operations in C++ (see Table 9.6) that combine C++ scripting at the R command line with formal C++ grammar, including header files.
src <-
'
#include <Rcpp.h>
#include <math.h>
using namespace Rcpp;
// [[Rcpp::export]]
List math_demo(){
double a = sin(3);
double b = log(3);
double c = log2(3);
NumericVector v = {1,2,3};
double d = min(v);
NumericVector e = log(v);
return List::create(Named("a") = a,
Named("b") = b,
Named("c") = c,
Named("d") = d,
Named("e") = e);
}'
sourceCpp(code = src)
math_demo()$a
[1] 0.14112
$b
[1] 1.0986
$c
[1] 1.585
$d
[1] 1
$e
[1] 0.00000 0.69315 1.09861\(\blacksquare\)
- The entire C++ script (Lines 2-21) is written into a character string, and assigned the name
src. - The first lines of C++ code include calls to both the
Rcpp.handmath.hheader files (Lines 3-4), application of the Rcpp namespace (Line 6), and designation of// [[Rcpp::export]](Line 7). - Lines 9-21 codify the C++ function
math_demo. The function is argumentless (it is meant to demonstrate mathematics using object generated in the function itself) and will return an RcppList(Line 9). - Lines 10-12 are simple scalar operations using
math.hfunctions. - Lines 13-15 use Rcpp
Vectorapproaches. - A
Listcontaining the generated objects,a,b,c,d, andeis built and returned on Lines 16-20.
9.5.1.6 Accessing/Manipulating Data Type Components
Rcpp data type objects can generally be subset using (), [], or with member functions. Both () and [] can be used with Rccp::NumericVector, Rcpp::IntegerVector and CharacterVector types. Rcpp::Dataframe objects require [], whereas Rcpp::Matrix, require () for subsetting.
Example 9.26 \(\text{}\)
Here is a long-winded C++ function that demonstrates Rcpp subsetting using Rcpp::NumericVector objects, an Rcpp::NumericMatrix object, and an Rcpp::Dataframe object.
#include <Rcpp.h>
using namespace Rcpp;
// [[Rcpp::export]]
List subsets(){
// Create Vectors
NumericVector nv = {10, 20, 30, 40, 50, 60};
nv.names() = CharacterVector({"A","B","C","D","E","F"});
NumericVector nv2 = nv + 1;
NumericVector nv3 = nv + nv2; // Rcpp allow elementwise Vector operations
// Create Matrix
NumericMatrix nm(2, 3, nv.begin());
// Create Dataframe
DataFrame df = DataFrame::create(Named("V2") = nv2, Named("V3") = nv3);
// Indexes
NumericVector id1 = {1,3};
CharacterVector id2 = {"A","D","E"};
LogicalVector id3 = {false, true, true, true, false, true};
// Vector subsets based on indexes
int x1 = nv[0];
int x2 = nv["C"];
NumericVector x3 = nv[id1];
NumericVector x4 = nv[id2];
NumericVector x5 = nv[id3];
// Matrix subsets
double x6 = nm(0 , 1); // Row 0 (first row) and column 1 (2nd column)
NumericVector x7 = nm(1 , _ ); // Row 1 (2nd row)
NumericVector x8 = nm( _ , 0); // Column 0 (1st column)
NumericVector x9 = nm.column(0); // Column 0 (1st column)
//Dataframe subsets
NumericVector x10 = df[0];
NumericVector x11 = df["V3"];
return List::create(Named("Result1") = x1, Named("Result2") = x2,
Named("Result3") = x3, Named("Result4") = x4,
Named("Result5") = x5, Named("Result6") = x6,
Named("Result7") = x7, Named("Result8") = x8,
Named("Result9") = x9, Named("Result10") = x10,
Named("Result11") = x11);
}
subsets()$Result1
[1] 10
$Result2
[1] 30
$Result3
B D
20 40
$Result4
A D E
10 40 50
$Result5
B C D F
20 30 40 60
$Result6
[1] 30
$Result7
[1] 20 40 60
$Result8
[1] 10 20
$Result9
[1] 10 20
$Result10
[1] 11 21 31 41 51 61
$Result11
[1] 21 41 61 81 101 121- As before, I call the
Rcpp.hheader file, apply the Rcpp namespace, and designate the attribute// [[Rcpp::export]](Lines 1-3). - On Line 4, the C++ function
subsetsis defined to haveListoutput. No arguments are defined because the goal is to demonstrate Rcpp data type subsetting and manipulation, using only objects created within the function. - On Lines 6-9, I create three
NumericVectorobjects. The latter two are on elementwise transformations facilitated by Rcpp sugar operators. - On Line 11, I create a
NumericMatrixfilled with elements from theVectornv, using theMatrixdeque member functionbegin(). Note that Rcpp matrices are built by column, given a vector input. - On Line 13, I create a two column
Dataframecomprised of theVectorobjectsnv2andnv3. using theMatrixdeque member functionbegin(). - On Line 15-17, three
Vectorobjects that will be used for subsequent subsetting are created. - On Lines 19-23, the objects
x1,x2,x3,x4andx5are created by subsetting theVector,nv. - On Lines 25-28, the objects
x6,x7,x8, andx9are created by subsetting theMatrix,nm. - On Lines 30-31, the objects
x10andx11are created by subsetting theDataframe,df. - On Lines 33-38, the subset objects are assembled into a
Listand are returned by the function.
\(\blacksquare\)
Example 9.27 \(\text{}\)
We now know enough to extend our scalar function for Simpson’s diversity (Example 9.24) to a function that can handle matrices –the conventional format for biological community datasets.
#include <Rcpp.h>
using namespace Rcpp;
// [[Rcpp::export]]
NumericVector simpson(NumericMatrix x) {
CharacterVector rn = rownames(x);
NumericVector out = x.nrow();
out.names() = rn;
int n = out.size();
for(int i = 0; i < n; ++i) {
NumericVector temp = na_omit(x(i , _ ));
double total = sum(temp);
NumericVector relsq = pow(temp/total, 2);
out[i] = 1 - sum(relsq);
}
return out;
}- As in previous examples, I first call the
Rcpp.hheader file, apply the Rcpp namespace, and define the// [[Rcpp::export]]compiler attribute (Lines 1-3). - The function output will be a
NumericVector(of Simpson’s diversities of sites) and will require aNumericMatrixfor its argumentx, with sites in rows and species in columns (Line 4). - Lines 5-8 generate objects (
outandn) that will be used in a subsequent loop. - Lines 10-15 define a loop that populates
outwith Simpson’s diversities. The code:NumericVector temp = na_omit(x(i , _ ));creates aNumericVectorobject,temp, consisting of non-missing values in the \(i\)th row ofx. - On Line 17
outis returned.
Here we apply our function to the entire vegan::varespec dataset.
simpson(as.matrix(varespec)) 18 15 24 27 23 19 22 16 28
0.82171 0.76276 0.78101 0.74414 0.84108 0.81819 0.80310 0.82477 0.55996
13 14 20 25 7 5 6 3 4
0.81828 0.82994 0.84615 0.83991 0.70115 0.56149 0.73888 0.64181 0.78261
2 9 12 10 11 21
0.55011 0.49614 0.67568 0.50261 0.80463 0.85896 We see that our C++ function is much faster than the widely-used function vegan::diversity(), which relies on an R for loop.
m <- matrix(nrow = 10^6, ncol = 10, data = rnorm(10^7) + 10)
system.time(simpson(m)) user system elapsed
0.30 0.04 0.36
system.time(vegan::diversity(m, "simpson")) user system elapsed
2.66 0.18 2.94 \(\blacksquare\)
9.5.2 The inline package
The inline R package (Sklyar, Eddelbuettel, and Francois 2025) extends the capacities of Rcpp::evalCpp(), Rcpp::cppFunction() and Rcpp::sourceCpp() by allowing users to create, compile, and run functions written in any language supported by R CMD SHLIB, including C, Fortran, C++, and C-obj, from the R command line.
Example 9.28 \(\text{}\)
Consider the following example –based on ? inline::cfunction()– of a simple C function that raises every value in a numeric vector to the third power.
library(inline)
code.cube <- "
int i;
for (i = 0; i < *n; i++)
x[i] = x[i]*x[i]*x[i];
"
cube.fn <- cfunction(signature(n="integer", x="numeric"), code.cube,
language = "C", convention = ".C")
cube.fn(20, 1:20)$n
[1] 20
$x
[1] 1 8 27 64 125 216 343 512 729 1000 1331 1728 2197 2744
[15] 3375 4096 4913 5832 6859 8000Note that code.cube, is a character string containing C-script (Lines 3-7). The script on Lines 9 and 10 calls inline::cfunction() to compile the string into a C shared library executable using SHILB. The shared library will be called automatically using .C(), allowing cube.fn used as an R function on Line 9. The object cube.fn has an unusual combination of characteristics. It is a function of base type closure:
typeof(cube.fn)[1] "closure"However, it is also S4,
isS4(cube.fn)[1] TRUEwith the following slots:
slotNames(cube.fn)[1] ".Data" "code" The code slot can be obtained using the function inline::code()
code(cube.fn) 1: #include <R.h>
2:
3:
4: void file3c6c2fcb2f1c ( int * n, double * x ) {
5:
6: int i;
7: for (i = 0; i < *n; i++)
8: x[i] = x[i]*x[i]*x[i];
9:
10: }Note that the text string has been converted to a C void function, as required by SHLIB. The header call #include <R.h> provides a built-in R API for C code.
The .Data slot contains R code that is run by interpreter when cube.fn() is called. Note that the function uses .Primitive() to call the appropriate shared library by way of its object address/pointer.
cube.fn@.Datafunction (n, x)
.Primitive(".C")(<pointer: 0x00007ffa2be01380>, n = as.integer(n),
x = as.double(x))
<environment: 0x0000023c47e45510>\(\blacksquare\)
9.6 SQL and Databases
Biological datasets have grown exponentially in both size and number (Sima et al. 2019). Because of this trend, biological databases are often housed in web-accessible warehouses, including the National Center for Biotechnology Information (NCBI), dataBase for Gene Expression Evolution (Bgee), and the European life-sciences infrastructure for biological information (ELIXIR). The Posit website provides a resource for working with databases in R.
Databases are often assembled in a Database Management System (DBMS) format. A DBMS will contain one or more rectangular row/column storage units called tables. Rows in tables are called records and columns are called fields or attributes.
Many DBMS formats have evolved based on the Structured Query Language (SQL). Although SQL is an American National Standards Institute (ANSI) and International Organization for Standardization (ISO) standard, there are many variants, and software for managing these languages is often proprietary (e.g., Oracle, Microsoft SQL Server) and potentially expensive. Despite this variety, SQL dialects generally use the same basic SQL commands (Table 9.8), and processes. For example, as a general rule, SQL table fields can be accessed with a period operator. That is, a column, bar, in table foo is specified as foo.bar. Most SQL variants require ending program instructions with a semicolon, ;.
SQL guidance can be found at a large number of websites, including the developer site W3.
| Command | Meaning |
|---|---|
SELECT |
Extracts data from a database |
FROM |
Used with SELECT. A clause identifying a database |
UPDATE |
Updates data in a database |
DELETE |
Deletes data from a database |
CREATE TABLE |
Creates a new table |
DROP TABLE |
Permanently remove a table |
INSERT INTO |
Add a new record to table |
VALUES |
Used with INSERT INTO define record values |
WHERE |
Filters records from a table |
AND |
Filters records based on more than one condition |
OR |
Filters records based on more than one condition |
BETWEEN |
Selects values within a given range |
9.6.1 DBI
The R package DBI (R Special Interest Group on Databases (R-SIG-DB), Wickham, and Müller 2024; James 2009) currently allows communication with 30 SQL-driven DBMS formats. Each supported DBI DBMS uses its own R package. For instance, the SQLite DBMS is interfaced with the package RSQLite (which will be installed with DBI), and the MySQL DBMS can be interfaced using the package RMySQL. The RMariaDB package can be used to interface either MySQL or the DBMS MariaDB. Opening a DBMS connection will constrain users to the SQL nuances of the selected DBMS. We will concentrate initially on the non-proprietary DBMS SQLite.
9.6.2 SQLite
SQLite and other SQL DBMS variants can generally be run directly from system shells, following their installation.
Example 9.29 \(\text{}\)
SQLite version 3.45.1 2024-01-30 16:01:20
Enter ".help" for usage hints.
Connected to a transient in-memory database.
Use ".open FILENAME" to reopen on a persistent database.
sqlite>Note that the command line prompt for the SQLite shell is: sqlite>.
To create a database called a_acid with a table called tbl1, containing amino acid information, I could do something like:
sqlite> CREATE TABLE tbl1(Name TEXT, Mass DOUBLE PRECISION);
sqlite> INSERT INTO tbl1 VALUES('Alanine', 89.094);
sqlite> INSERT INTO tbl1 VALUES('Aspartine', 174.203 );
sqlite> .mode table
sqlite> SELECT * FROM tbl1;+-----------+---------+
| Name | Mass |
+-----------+---------+
| Alanine | 89.094 |
| Aspartine | 174.203 |
+-----------+---------+- On Line 1, I open (and simultaneously create in my home directory) the database
a_acid_db, as I open SQLite. - On Line 2, I create
tbl1which will have two fields. The first will contain amino acid name entries which will be store as text. The second will contain molecular weights, stored as double precision numbers. - On Lines 3 and 4 I add records for the table.
- On Line 5, I define the
.modethat I want SQLite to use when printing/summarizing queries. Other.modeoptions include.mode list,.mode line, and.mode box. - On Line 6, the command
SELECT * FROM tbl1means “select and show” all columns fromtbl1.
Typing .exit or .quit will close both the current SQLite shell session and any open database(s). connections.
Opening an existing database at the SQLite command line requires the function .open.
The function .tables allows one to view tables within an open database.
tbl1\(\blacksquare\)
9.6.3 RSQLite
To demonstrate interfacing SQLite with R, I will create a database using existing “internal” R dataframes.
Example 9.30 \(\text{}\)
First, we load DBI and RSQLite.
Next, we establish a SQLite DBMS connection using dbConnect().
<SQLiteConnection>
Path: :memory:
Extensions: TRUEUnlike many other DBMS frameworks that may require a username, password, host, port, and other information, SQLite only requires a path to the database127. The argument ":memory:" specifies a special path that results in an “in-memory” database.
Notably, the con database is an S4 object:
isS4(con)[1] TRUEHere we append the asbio::world.emission dataframe to the database using dbWriteTable().
library(asbio)
data(world.emissions)
dbWriteTable(con, "emissions", world.emissions)We see that the table (renamed emissions) now exists in the database.
dbListTables(con)[1] "emissions"Below we use SQL script (within the R function DBI::dbSendQuery()) to access information from the database table emissions. In particular –using the commands SELECT, FROM, WHERE, AND, and BETWEEN– I query the columns coal_co2 and gas_co2, with respect to the United States, for the years 2016 to 2019.
us <- dbSendQuery(con, "SELECT coal_co2, gas_co2
FROM emissions
WHERE country = 'United States'
AND year BETWEEN 2016 AND 2019")Recall (Example 9.29) that to access all columns from emissions, I could have used the SQL command: "SELECT * FROM emissions.
Here I fetch the query result using dbFetch():
us.fetch <- dbFetch(us)
us.fetch coal_co2 gas_co2
1 1378.2 1509.0
2 1337.5 1491.8
3 1282.1 1653.0
4 1094.7 1706.9The fetched result is a dataframe.
class(us.fetch)[1] "data.frame"One should clear queries using DBI::dbClearResult(). This will free all computational resources (local and remote) associated with the RSQLite query result.
dbClearResult(us)Databases can contain multiple tables. Here I append the asbio::C.isotope dataframe to the database:
data(C.isotope)
dbWriteTable(con, "isotopes", C.isotope)There are now two tables in the database, although they are not relational.
dbListTables(con)[1] "emissions" "isotopes" When finished accessing a DBMS, one should always close the DBMS connection.
dbDisconnect(con)\(\blacksquare\)
One can use the SQL variant of the chosen DBMS directly in a knitr chunk by specifying the options ```{sql, connection = con}```, where con is the name of the database connection (see Section 9.1.4). This approach will often be required for complex SQL operations in R.
Example 9.31 \(\text{}\)
Reconsidering Example 9.30 we have:
con <- dbConnect(SQLite(), ":memory:")
dbWriteTable(con, "emissions", world.emissions)Here I directly specify an SQL query (in SQL).
SELECT coal_co2, gas_co2
FROM emissions
WHERE country = 'United States'
AND year BETWEEN 2016 AND 2019;| coal_co2 | gas_co2 |
|---|---|
| 1378.2 | 1509.0 |
| 1337.5 | 1491.8 |
| 1282.1 | 1653.0 |
| 1094.7 | 1706.9 |
dbDisconnect(con)\(\blacksquare\)
9.6.4 Relational DBMS
Thus far, the justification for an interfaced DBMS may seem vague, since similar data management results could be obtained from R lists.
The advantages of creating a DBMS become clearer when considering a relational DBMS (RDBMS) framework. An RDBMS allows the straightforward linking of multiple database tables via a common value identifier stored in the tables (Fig 9.2).
. Several tables are linked via the identifier **SpeciesID**. Figure taken from @sima2019.](figs3/relational.png)
FIGURE 9.2: A relational database from the gene expression database Bgee. Several tables are linked via the identifier SpeciesID. Figure taken from Sima et al. (2019).
Example 9.32 \(\text{}\)
In this example we will impart relational characteristics to a database based on two R dataframes, asbio::Rabino_CO2 and asbio::Rabino_del13C, obtained from (Rubino et al. 2013). The datasets record CO\(_2\) and \(\delta^{13}\)C levels from Law Dome and South Pole, Antarctica for a 1000 year timespan. Exact effective date records, precision, and measurement depths all vary for the entries (see Example 7.5), prompting the creation of two separate datasets.
First, I create mean effective date records to eventually provide a single-entry label field for each dataset, based on the effective.age of samples.
data(Rabino_CO2)
data(Rabino_del13C)
library(tidyverse)
AvgCO2df <- Rabino_CO2 |>
group_by(effective.age) |>
summarise(AvgDdepth = mean(depth),
AvgCO2 = mean(CO2),
AvgUncertainty = mean(uncertainty))
Avg13Cdf <- Rabino_del13C |>
group_by(effective.age) |>
summarise(AvgDepth = mean(depth),
Avgd13C = mean(d13C.CO2),
AvgUncertainty = mean(uncertainty))
names(Avg13Cdf)[1] <- names(AvgCO2df)[1] <- "EffectiveAge"
AvgCO2df$EffectiveAge <- as.integer(unlist(AvgCO2df[,1]))
Avg13Cdf$EffectiveAge <- as.integer(unlist(Avg13Cdf[,1]))The resulting summary dataframes, AvgCO2df and AvgC13df, do not contain measures from the same effective dates. Specifically, 114 (out of 189) AvgCO2df effective age records do not occur in AvgC13df.
[1] 114And 10 (out of 85) AvgC13df effective age records do not occur in AvgCO2df.
[1] 10Nonetheless, we can easily join the datasets in a DBMS, and use their effective ages, to simultaneously query them.
We first request a SQLite database connection.
We then add AvgCO2df and AvgC13df to the database as tables.
dbWriteTable(con, "CO2", AvgCO2df)
dbWriteTable(con, "d13C", Avg13Cdf)There are several database joins we can specify using SQL, including LEFT JOIN and RIGHT_JOIN. Assume that we have two tables in a database , A and B.
If I request A LEFT JOIN B, then the result set will include:
- Records in
AandBwith corresponding labels. - Records (if any) in
Awithout corresponding labels inB. In this case,Bentries are givenNAvalues.
Conversely, if I request A RIGHT JOIN B, then the result set will include:
- Records in
BandAwith corresponding labels. - Records (if any) in
Bwithout corresponding labels inA. In this case,Aentries are givenNULLvalues.
SELECT AvgCO2, d13C.Avgd13C, CO2.EffectiveAge
FROM CO2 LEFT JOIN d13C
ON d13C.EffectiveAge = CO2.EffectiveAge
WHERE CO2.EffectiveAge > 1990;| AvgCO2 | Avgd13C | EffectiveAge |
|---|---|---|
| 352.22 | -7.8410 | 1991 |
| 353.73 | -7.8820 | 1992 |
| 353.94 | -7.8883 | 1993 |
| 357.11 | NA | 1994 |
| 359.65 | NA | 1996 |
| 361.78 | -8.0600 | 1998 |
| 368.02 | -8.0695 | 2001 |
In the SQL code above, I specify a LEFT JOIN.
- On Line 1, I specify the fields whose data I want to consider jointly,
AvgCO2,d13C.Avgd13C, and the reference field I wish to use,CO2.EffectiveAge, i.e., theEffectiveAgefield in theCO2table. - On Line 2, I specify the join:
CO2 LEFT JOIN d13C. - On Line 3, I identify the fields used to join the tables.
- On Line 4, I limit the printed results to
CO2.EffectiveAgevalues greater than 1990.
Note that in the output above there are two effective ages, 1994 and 1996, with CO\(_2\) records but no \(\delta^{13}\)C records.
SELECT AvgCO2, d13C.Avgd13C, CO2.EffectiveAge
FROM CO2 RIGHT JOIN d13C
ON d13C.EffectiveAge = CO2.EffectiveAge
WHERE CO2.EffectiveAge > 1990;| AvgCO2 | Avgd13C | EffectiveAge |
|---|---|---|
| 352.22 | -7.8410 | 1991 |
| 353.73 | -7.8820 | 1992 |
| 353.94 | -7.8883 | 1993 |
| 361.78 | -8.0600 | 1998 |
| 368.02 | -8.0695 | 2001 |
The RIGHT JOIN SQL statement above is identical to the previous statement except for the Line 2 command: CO2 RIGHT JOIN d13C. In the output, complete \(\delta^{13}\)C records for the requested effective age range are returned (note that ages 1994 and 1996 are omitted). While not required by the query, corresponding records for CO\(_2\) also exist, and are reported.
dbDisconnect(con)\(\blacksquare\)
9.6.5 Creating an SQLite Relational Database
Thus far we have used R package dataframes to populate an SQLite database connection. An important application is saving an SQLite database from related but intentionally separated data files like those considered in Example 9.32.
Example 9.33 \(\text{}\)
Here I open a database to contain the dataframes AvgCO2df and Avg13Cdf (as created in Example 9.32), then save the result under the name C_db.sqlite in my working directory.
con <- dbConnect(SQLite(), ":memory:")
dbWriteTable(con, "CO2", AvgCO2df)
dbWriteTable(con, "d13C", Avg13Cdf)
sqliteCopyDatabase(con, "C_db.sqlite")
dbDisconnect(con)To verify the contents of the C_db.sqlite database, I close the database connection open C_db.sqlite using RSQLite::dbConnect.
dbListTables(con)[1] "CO2" "d13C"
dbDisconnect(con)\(\blacksquare\)
9.7 Python
Python, whose image logo is shown in Fig 9.3, is similar to R in several respects. Python was formally introduced in the early 90s, is an open source OOP language that is rapidly gaining popularity, and its source code is usually evaluated in an on-the-fly manner. That is Python, like R, is generally used as an interpreted language. Like R, comments in Python are made using the pound metacharacter, #128, and many function calls have similar syntax.

FIGURE 9.3: The symbol for Python, a high-level, general-purpose, programming language.
There are, however, several fundamental differences between Python and R. These include the fact that while white spaces in R code (including tabs) simply reflect coding style preferences –for example, to increase code clarity– Python indentations denote code blocks129. That is, Python indentations serve the same purpose as R curly braces. Another important difference is that R object names can contain a . (dot), whereas in Python . means: “attribute in a namespace.” Thus, in Python the period operator . serves the same role as $ in R lists, dataframes and environments (Sections 3.1.4, 8.8.1.1). Recall that the . operator is used in a similar way in SQL language queries of database tables (Section 9.6). Python also uses member functions (Section 9.5.1.2) for class methods instead of generic function calls like R. In Python, a method for an object of a specific class is specified using the period operator. Useful guidance for converting R code to analogous Python code can be found here.
Python can be downloaded for free from (https://www.python.org/downloads/), and can be activated from the Windows shells using the commands py or python, and activated from Mac and Unix/Linux shells using the command python. General guidance for the Python language can be found at (https://docs.python.org/) and many other sources including these books. Below I call Python from the Windows PowerShell command line.
Python 3.13.7 (tags/v3.13.7:bcee1c3, Aug 14 2025, 14:15:11) [MSC v.1944 64 bit (AMD64)] on win32
Type "help", "copyright", "credits" or "license" for more information.
>>>Note that the standard command line prompt for the Python shell is >>>. We can exit Python from the command line by typing quit().
9.7.1 reticulate
Module(numpy)Because our primary interest is interfacing Python and R, and not Python itself, we will use R as our base of operations. This will require the R package reticulate (Ushey, Allaire, and Tang 2023).
# install.packages("reticulate")
library(reticulate)RStudio (via reticulate) can be used as an IDE for Python130. In this capacity RStudio will:
- Generate a Python-specific environment (to provide separate settings for Python and R objects).
- Call separate R and Python environments, depending on which language is currently used in a chunk. Python code can be run directly from knitr chunks by defining
python(instead ofr) as the first chunk option (Section 9.1.4).
9.7.2 Important Considerations for IDEs and APIs
Python packages are currently installed from one of two package repositories: the Python Package Index (PyPI) or Anaconda. The Python Installer Package (pip) is designed to install packages from PyPI (see Section 9.7.3). A repository manager named conda is used in conjunction with Anaconda, and its stripped-down repository Miniconda.
Using Python can be a headache if: 1) different versions of Python exist on your machine, and it is unclear which versions (if any) have access to necessary repositories131, and/or 2) Python installations and packages are accessible under one manager (e.g., pip), but not another (e.g., conda). This is further complicated by the fact that Python APIs (like reticulate) or IDEs (like Spyder) may come with their own repository frameworks and default versions of Python.
With this mind, I can specify a path to a specific Python executable to be used in an R/reticulate session with reticulate::use_python(), and specify a repository path (and a path a particular Python executable) with reticulate::use_condaenv() and reticulate::use_miniconda(). The code below specifies use of my (external to reticulate) Miniconda environment as a package repository and Python executable path.
use_condaenv("C:/Users/ahoken/miniconda3/")
The version of Python used by reticulate can be accessed with Sys.which(), which finds full paths to program executables.
Sys.which("python") python
"C:\\Users\\ahoken\\MINICO~1\\python.exe" More details concerning my Python configuration for reticulate are revealed with reticulate::py_config():
reticulate::py_config()python: C:/Users/ahoken/miniconda3/python.exe
libpython: C:/Users/ahoken/miniconda3/python313.dll
pythonhome: C:/Users/ahoken/miniconda3
version: 3.13.5 | packaged by Anaconda, Inc. | (main, Jun 12 2025, 16:37:03) [MSC v.1929 64 bit (AMD64)]
Architecture: 64bit
numpy: C:/Users/ahoken/miniconda3/Lib/site-packages/numpy
numpy_version: 2.3.1
numpy: C:\Users\ahoken\MINICO~1\Lib\site-packages\numpy
NOTE: Python version was forced by use_python() functionA Python command line REPL (Read-Eval-Print Loop) interface can be called directly in R using:
reticulate::repl_python()Python can be closed from the resulting interface (returning one to R) by typing:
Example 9.34 \(\text{}\)
The following are Python operations, run directly from RStudio.
4The Python assignment operator is =.
4Here we see the aforementioned importance of indentation.
positiveLack of an indented “block” following if will produce an error. Indentations in code can be made flexibly (e.g., one space, two space, tab, etc.) but they should be used consistently.
\(\blacksquare\)
9.7.3 Packages
Like R, Python consists of a core language, a set of built-in functions, modules, and libraries (i.e., the Python standard library), and a vast collection (\(>200,000\)) of supplemental libraries. Imported libraries are extremely important in Python because its distributed version has limited functional capabilities (compared to R). A number of important Python supplemental libraries, each of which contain multiple packages, are shown in Table 9.9.
| Library | Purpose |
|---|---|
| sumpy | Fundamental package for scientific computing |
| scipy | Mathematical functions and routines |
| matplotlib | 2- and 3-dimensional plots |
| pandas | Data manipulation and analysis |
| sympy | Symbolic mathematics |
| bokeh | Interactive data visualizations |
We can install Python packages and libraries using the pip package manager for Python or conda (Section 9.7.2). Installation only needs to occur once on a workstation (similar to install.packages() in R). Following installation, one can load a package for a particular work session using the Python function import (analogous to library() in R)132.
Installation of a Python package, foo, with reticulate, can be accomplished using the function reticulate::py_install (in R)133.
py_install("foo")Example 9.35 \(\text{}\)
I wish to install the ecologits library, and its dependencies in the openAI library. This requires use of pip, via conda. Hence, I use the command:
py_install("ecologits[openai]", method = "conda", pip = TRUE) # Run in RTo load the ecologits library I use the Python function import():
\(\blacksquare\)
9.7.4 Functions in Packages
Functions within Python packages are obtained using a package.function syntax. Here I import numpy and run the function pi (which is contained in numpy).
3.141592653589793If we are writing a lot of numpy functions, Python will allow you to define a simplified library prefix. For instance, here I created a shortcut for numpy called np and use this shortcut to access the numpy functions pi() and sin().
np.float64(0.3420201433256687)
Use of the command from numpy import * would cause names of functions from NumPy to overwrite functions with the same name from other packages. That is, we could run numpy.pi simply using pi.
Example 9.36 \(\text{}\)
Here we import the package pyplot from the library matplotlib, rename the package plt, and create a plot (Fig 9.4) using the function pyplot.plot() (as plt.plot()) by calling:
FIGURE 9.4: Creating a Python plot using R.
In Line 2, the command range(10) creates a sequence of integers from zero to ten. This is used as the first argument of plt.plot(), which specifies the plot \(x\)-coordinates. If \(y\) coordinates are not specified in the second argument, \(x\)-coordinates will be reused as \(y\) coordinates. The command 'bo' places blue filled circles at \(x\),\(y\) coordinates. Documentation for matplotlib.pyplot.plot() can be found at the matplotlib.org website.
\(\blacksquare\)
9.7.5 Data Types
There are four major built-in dataset storage classes in Python: lists, tuples, sets, and dictionaries (Table 9.10).
| Storage type | Example | Entry characteristics |
|---|---|---|
| List | ["hot","cold"] |
Changeable, Duplicates OK |
| Tuple | ("hot","cold") |
Unchangeable, Duplicates OK |
| Set | {"hot","cold"} |
Unchangeable, Duplicates not OK |
| Dictionary | {"temp":["hot", cold"]} |
Changeable, Duplicates not OK |
All four classes track element order and can be used to simultaneously store different types of data, e.g., character string and numbers.
We can make a Python list, which can contain both text and numeric data, using square brackets or the function list().
Classes of Python objects can be identified with the Python function type().
<class 'list'>An empty list can be specified as []
[]Like R, we can index list elements using square brackets. Importantly, like C-alike languages, indices start at 0. That is, a[0] refers to the first element of the list a.
20And the third element would be:
'Hi'As with R, square brackets can also be used to reassign list values
[20, 7, 'Hi', 10, 'end']We can use the function append() to append entries to the end of a list. For instance, to append the number 9 to the object a in the previous example, I would type:
[20, 7, 'Hi', 10, 'end', 9]The function appendleft() can be used to efficiently append entries to the beginning of an object of class deque (from the Python collections package). The function deque() can be used to convert a list into a deque (double ended queue).
<class 'collections.deque'>deque([0, 20, 7, 'Hi', 10, 'end', 9]) Unlike a Python list, a data object called a tuple, which is delineated using parentheses, contains elements that cannot be changed:
1Multidimensional numerical arrays, including matrices, can be created using functions from numpy.
Example 9.37 \(\text{}\)
Here we define: \[\boldsymbol{B} =
\begin{bmatrix}
1 & 4 & -5 \\
9 & -7.2 & 4
\end{bmatrix}\]
array([[ 1. , 4. , -5. ],
[ 9. , -7.2, 4. ]])We see that B is an object of class numpy.ndarray (meaning numpy n-dimensional array).
<class 'numpy.ndarray'>Mathematical matrix operations can be easily applied to numpy.ndarray objects. Here I find \(\boldsymbol{B} - 5\)
array([[ -4. , -1. , -10. ],
[ 4. , -12.2, -1. ]])Extensive linear algebra tools are contained in the libraries numpy and scipy.
\(\blacksquare\)
Unlike a list, a numpy array will allow Boolean indexing and vectorized operations.
9.7.6 Member Functions and Other Attributes
Like C++, Python uses a member function approach to create and call methods for particular classes. As with C++, a member function foo, for a class underlying an object bar, would be run as: bar.foo(). Python classes often have special member functions called magic methods or dunder (short for double underline) methods . These would be called using the syntax: bar.__foo__(). Python also uses instance variables which are automatically stored as a data attribute of a particular class, but do not require a methods call. An instance variable foo for an object bar would be called using: bar.foo. Available methods and instance variables for an object bar can be listed using dir(bar), or `bar.__dir__(), assuming the object has a .__dir__() dunder method.
Example 9.38 \(\text{}\)
Consider the numpy.ndarray object B from Example 9.37. There are a large number of object attributes.
['T', '__abs__', '__add__', '__and__', '__array__', '__array_finalize__', '__array_function__', '__array_interface__', '__array_namespace__', '__array_priority__', '__array_struct__', '__array_ufunc__', '__array_wrap__', '__bool__', '__buffer__', '__class__', '__class_getitem__', '__complex__', '__contains__', '__copy__', '__deepcopy__', '__delattr__', '__delitem__', '__dir__', '__divmod__', '__dlpack__', '__dlpack_device__', '__doc__', '__eq__', '__float__', '__floordiv__', '__format__', '__ge__', '__getattribute__', '__getitem__', '__getstate__', '__gt__', '__hash__', '__iadd__', '__iand__', '__ifloordiv__', '__ilshift__', '__imatmul__', '__imod__', '__imul__', '__index__', '__init__', '__init_subclass__', '__int__', '__invert__', '__ior__', '__ipow__', '__irshift__', '__isub__', '__iter__', '__itruediv__', '__ixor__', '__le__', '__len__', '__lshift__', '__lt__', '__matmul__', '__mod__', '__mul__', '__ne__', '__neg__', '__new__', '__or__', '__pos__', '__pow__', '__radd__', '__rand__', '__rdivmod__', '__reduce__', '__reduce_ex__', '__repr__', '__rfloordiv__', '__rlshift__', '__rmatmul__', '__rmod__', '__rmul__', '__ror__', '__rpow__', '__rrshift__', '__rshift__', '__rsub__', '__rtruediv__', '__rxor__', '__setattr__', '__setitem__', '__setstate__', '__sizeof__', '__str__', '__sub__', '__subclasshook__', '__truediv__', '__xor__', 'all', 'any', 'argmax', 'argmin', 'argpartition', 'argsort', 'astype', 'base', 'byteswap', 'choose', 'clip', 'compress', 'conj', 'conjugate', 'copy', 'ctypes', 'cumprod', 'cumsum', 'data', 'device', 'diagonal', 'dot', 'dtype', 'dump', 'dumps', 'fill', 'flags', 'flat', 'flatten', 'getfield', 'imag', 'item', 'itemset', 'itemsize', 'mT', 'max', 'mean', 'min', 'nbytes', 'ndim', 'newbyteorder', 'nonzero', 'partition', 'prod', 'ptp', 'put', 'ravel', 'real', 'repeat', 'reshape', 'resize', 'round', 'searchsorted', 'setfield', 'setflags', 'shape', 'size', 'sort', 'squeeze', 'std', 'strides', 'sum', 'swapaxes', 'take', 'to_device', 'tobytes', 'tofile', 'tolist', 'trace', 'transpose', 'var', 'view']Note that dunder methods are listed first, and conventional member functions and instance variables are grouped together at the end of the dir() output.
The dunder method .__abs__() prints the elementwise absolute values of the array and .__len__() gives the number of rows.
array([[1. , 4. , 5. ],
[9. , 7.2, 4. ]])2The instance variables for an array include .shape (which reports the number of rows, columns, etc.) and .size which returns the number of array elements.
(2, 3)6One can easily easily obtain the mean and standard deviation of any array using the array member functions .mean() and .std().
np.float64(0.9666666666666667)np.float64(5.556277730839435)\(\blacksquare\)
9.7.7 Boolean Operations
Python Boolean operators are very similar to those in R (Table 9.11). As exceptions, to designate “and” and “or” in Python, one would use the commands and and or, respectively. Additionally, Python (like C and C++) uses True and False, instead of TRUE and FALSE.
| Operator | Operation | To ask: | We type: |
|---|---|---|---|
> |
\(>\) | Is x greater than y? |
x > y |
>= |
\(\geq\) | Is x greater than or equal to y? |
x >= y |
< |
\(<\) | Is x less than y? |
x < y |
<= |
\(\leq\) | Is x less than or equal to y
|
x <= y |
== |
\(=\) | Is x equal to y? |
x == y |
!= |
\(\neq\) | Is x not equal to y? |
x != y |
and |
and | Do x and y equal z? |
x == z and y == z |
& |
and (bitwise) | Bitwise comparison of x and y
|
x & y |
| or | or | Do x or y equal z? |
x == z or y == z |
| | | or (bitwise) | Bitwise comparison of x or y
|
x | y
|
Unlike R and C, && and || are not valid Boolean operators in Python.
Example 9.39 \(\text{}\)
Consider the following simple examples:
FalseTrueTrueFalseTrue\(\blacksquare\)
Unlike R, & and | are Python bitwise Boolean operators for “and” and “or”, respectively (Table 9.11). That is, they compare objects by paired bits (see Section 12.3) and, for each bit, return 1 for True and 0 for False.
Example 9.40 \(\text{}\)
This example will use the functions asbio::dec2bin() and asbio::bin2dec() to translate between binary {0, 1} and conventional (decimal) representations of numbers. For additional background see Sections 12.5 and 12.4.
The number 11 can be expressed in binary with four bits: 1011.
asbio::dec2bin(11)[1] 1011Whereas the number 14 can be expressed as: 1110.
asbio::dec2bin(14)[1] 1110In a bitwise comparison of the number 11 and 14, the first and third bits are equal (both equal 1) while the second and fourth bits are not equal. Thus, the bitwise Boolean result is 1010. This turns out to be the binary version of the number 10.
asbio::bin2dec(1010)[1] 10This result corresponds to the result of a bitwise comparison of the numbers 11 and 14 in Python, using &.
10\(\blacksquare\)
9.7.8 Mathematical Operations
Basic Python mathematical operators are generally (but not always) identical to R. For instance, note that for exponentiation ** is used instead of ^ (Table 9.12). This convention is also used by several other programming languages, including Fortran. Recallthat * can also +-be used by Python in non-mathematical contexts, for instance to load all function names from a package (Section 9.7.4).
| Operation | Operator/Function | To find | We type |
|---|---|---|---|
| addition | + |
\(2 + 2\) | 2 + 2 |
| subtraction | - |
\(2 - 2\) | 2 - 2 |
| multiplication | * |
\(2 \times 2\) | 2 * 2 |
| division | / |
\(\frac{2}{3}\) | 2/3 |
| modulo | % |
remainder of \(\frac{5}{2}\) | 5%2 |
| integer division | // |
\(\frac{5}{2}\) without remainder | 5//2 |
| exponentiation | ** |
\(2^3\) | 2**3 |
| \(\sqrt{x}\) | sqrt(x) |
\(\sqrt{2}\) | numpy.sqrt(2) |
| \(x!\) | factorial(x) |
\(5!\) | numpy.math.factorial(5) |
| \(\log_e\) | log(x) |
\(\log_e(3)\) | numpy.log(3) |
| \(e^x\) | exp(x) |
\(e^1 = 2.718282\dots\) | numpy.exp(1) |
| \(\pi = 3.141593 \dots\) | pi |
\(\pi\) | numpy.pi |
| \(\infty\) | inf |
\(\infty\) | float('inf') |
| \(-\infty\) | -inf |
\(-\infty\) | float('-inf') |
Symbolic derivative solutions to functions can be obtained using functions from the library sympy. Results from the package functions can be printed in LaTeX for pretty mathematics.
py_install("sympy", pip = TRUE) # run in R if sympy hasn't been installedExample 9.41 \(\text{}\)
Here we solve: \[\frac{d}{dx} 3e^{-x^2}\]
\[- 6 x e^{- x^{2}}\]
In Line 2, x is defined symbolically using the sympy.symbols() function. The variable x is used as a term in the expression fx in Line 3. The function fx is differentiated in Line 4 using the function sympy.diff().
\(\blacksquare\)
Integration in Python can be handled with the function quad() in scipy.
Example 9.42 \(\text{}\)
Here we find: \[\int_0^1 3e^{-x^2} dx\]
To perform integration we must install the scipy.integrate library using pip and bring in the function quad().
We then define the integrand as a Python function using the function def(). That is, def() is analogous to function() in R.
We now run quad() on the user function f with the defined bounds of integration.
(2.240472398437281, 2.487424042782217e-14)The first number is the value of the definite integral (in this case, the area under the function f from 0 to 1). The second is a measure of the absolute error in the numerical approximation.
\(\blacksquare\)
9.7.9 Reading in Data
Data in delimited files, including .csv files, can be read into Python using the numpy function loadtxt().
Example 9.43 \(\text{}\)
Assume that we have a comma separated dataset, named ffall.csv, located in the Python working directory, describing the free fall properties of some object over six seconds, with columns for observation number, time (in seconds), altitude (in mm) and uncertainty (in mm). The Python working directory (which need not be the same as the R working directory in RStudio) can be identified using the function getcwd() from the library os.
'C:\\Users\\ahoken\\Documents\\GitHub\\Amalgam'We can load freefall.csv using:
The first row was skipped (using skiprows = 1) because it contained column names and those were re-assigned when I brought in the data. Note that, unlike R, columns in the dataset are automatically attached to the global environment upon loading, and will overwrite objects with the same name.
array([0.18 , 0.182, 0.178, 0.165, 0.16 , 0.148, 0.136, 0.12 , 0.099,
0.083, 0.055, 0.035, 0.005])File readers in pandas are less clunky (and more similar to R). We can bring in freefall.csv using the function pandas.read_csv():
py_install("pandas") # Run if pandas is not installed obs time height error
0 1 0.0 180 3.50
1 2 0.5 182 4.50
2 3 1.0 178 4.00
3 4 1.5 165 5.50
4 5 2.0 160 2.50
5 6 2.5 148 3.00
6 7 3.0 136 2.50
7 8 3.5 120 3.00
8 9 4.0 99 4.00
9 10 4.5 83 2.50
10 11 5.0 55 3.60
11 12 5.5 35 1.75
12 13 6.0 5 0.75The object ffall is a Pandas DataFrame, which is different in several respects, from an R dataframe.
<class 'pandas.core.frame.DataFrame'> Column arrays in ffall can be called using the syntax: ffall., or by using braces, []. For instance:
0 180
1 182
2 178
3 165
4 160
5 148
6 136
7 120
8 99
9 83
10 55
11 35
12 5
Name: height, dtype: int640 180
1 182
2 178
3 165
4 160
5 148
6 136
7 120
8 99
9 83
10 55
11 35
12 5
Name: height, dtype: int64\(\blacksquare\)
9.7.10 Data Analysis in both Python and R
In RStudio, R and Python (reticulate) sessions are considered separately. When accessing Python from R, R data types are automatically converted to their equivalent Python types. Conversely, when values are returned from Python to R they are converted back to R types. It is possible, however, to access each from the others’ session.
The reticulate command py allows one to interact with a Python session directly from the R console. Here I convert the pandas DataFrame ffall into a recognizable R dataframe, within R.
ffallR <- py$ffallWhich allows me to examine it with R functions.
colMeans(ffallR) obs time height error
7.0000 3.0000 118.9231 3.1615 On Lines 1 and 2 in the chunk below, I bring in the Python library pandas (from R) with the function reticulate:import(). The code pd <- import("pandas", convert = FALSE) is the Python equivalent of: import pandas as pd.
pd <- import("pandas", convert = FALSE)As expected, the column names constitute the names attribute of the dataframe ffallR.
names(ffallR)[1] "obs" "time" "height" "error" The ffall dataframe, however, has different characteristics when it is loaded as a pandas DataFrame. Note that in the code below the pandas function read_csv() is accessed using pd$read_csv() instead of pd.read_csv() because an R chunk is being used.
ffallP <- pd$read_csv("ffall.csv")The names attribute of the pandas DataFrame ffallP, as perceived by R, contains over 200 entities due the presence of DataFrame attributes (including member functions and instance variables) see Section 9.7.6. Many of these are provided by the built-in Python module statistics. Here are the first 20.
[1] "abs" "add" "add_prefix" "add_suffix" "agg"
[6] "aggregate" "align" "all" "any" "apply"
[11] "applymap" "asfreq" "asof" "assign" "astype"
[16] "at" "at_time" "attrs" "axes" "backfill" I can call these attributes using the $ operator, in the style of RC and R6 methods (Section 8.7.2). These procedures clearly demonstrate the straightforwardness of R/Python syntheses under reticulate.
ffallP$mean()obs 7.000000
time 3.000000
height 118.923077
error 3.161538
dtype: float64
ffallP$var()obs 15.166667
time 3.791667
height 3495.243590
error 1.512147
dtype: float64
ffallP$kurt()obs -1.200000
time -1.200000
height -0.692166
error 0.445443
dtype: float64Note that the final result is clearly being provided by Python because kurtosis functions are not native to the R stats package.
For further analysis in R these attributes will need to be explicitly converted to R objects using the function py_to_r().
obs time height error
7.0000 3.0000 118.9231 3.1615 9.7.11 Python versus R
R generally allows much greater flexibility than Python for explicit statistical analyses and graphical summaries. For example, the Python statistics library Pymer4 actually uses generalized linear mixed effect model (see K. Aho (2014)) functions from the R package lme4 to complete computations. Additionally, Python tends to be less efficient than R for pseudo-random number generation134, since it requires looping to generate multiple pseudo-random outcomes (see Van Rossum and Drake (2009)).
Example 9.44 \(\text{}\)
Here I generate \(10^8\) pseudo-random outcomes from a continuous uniform distribution (processor details footnoted in Example 9.18).
R:
system.time(ranR <- runif(1e8)) user system elapsed
2.31 0.19 2.50 Python:
import time
import random
ranP = []
start_time = time.time()
for i in range(0,9999999):
n = random.random()
ranP.append(n)
time.time() - start_time3.0007407665252686The operation takes much longer for Python than R.
The Python code above requires some explanation. On Lines 1 and 2, the Python modules time and random are loaded from the Python standard library, and on Line 3 an empty list ranP is created that will be filled as the loop commences. On Line 5, the start time for the operation is recorded using the function time() from the module time. On Line 6 a sequence of length \(10^8\) is defined as a reference for the index variable i as the for loop commences. On Lines 7 and 8 a random number is generated using the function random() from the module random and this number is appended to ranP. Note that Lines 7 and 8 are indented to indicate that they reside in the loop. Finally, on Line 9 the start time is subtracted from the end time to get the system time for the operation.
\(\blacksquare\)
On the other hand, the system time efficiency of Python may be better than R for many applications, including the management of large datasets (Morandat et al. 2012).
Example 9.45 \(\text{}\)
Here I add the randomly generated dataset to itself in R:
system.time(ranR + ranR) user system elapsed
0.19 0.08 0.27 and Python:
0.11950039863586426For this operation, Python is faster.
\(\blacksquare\)
Of course, IDEs like RStudio allow, through the package reticulate, simultaneous use of both R and Python systems, allowing one to draw on the strengths of each language.
Exercises
- Complete the following exercises using either PowerShell or BASH system shells.
- Identify your home and root directory.
- List the contents of your home and root directories.
- Navigate to a directory on your computer with many files. List all files with a particular extension (e.g.,
.txt,.doc, etc.). - For the directory in (c), list all files and sort them alphabetically and by extension, using
sort. - Write your result from (d) to a text file using
>. - Use BASH (on Windows machines this will require installation of the Windows Subsystem for Linux). For the directory in (c), count the number of times the word
biologyoccurs in files usinggrep.
- Complete the following exercises using a BASH system shell. On Windows machines this will require installation of the Windows Subsystem for Linux.
- Create an empty shell program file named
dirsummary.shusing> dirsummary.sh(in BASH) orfile.create(dirsummary.sh)in R. - Open the file in nano and insert the code below and exit nano.
- Explain what is happening in each line of code.
- Create an empty shell program file named
#!/bin/bash
echo "The directories and files below are in: $(pwd) "
echo ""
ls -1 | head -n 8
echo ""
echo "The current time is: $(date) "- For Q. 2 above
- Set the file mode for
dirsummary.shin BASH usingchmod a+x dirsummary.sh. - Run the program using
./dirsummary.sh.
- Set the file mode for
- The Fortran script below calculates the circumference of the earth (in km) for a given latitude (measured in radians). For additional information, see Question 6 from the Exercises in Ch 2. Explain what is happening in each line of code.
Create a file
circumf.f90containing the code above and save it to an appropriate directory. Take a screen shot of the directory.Compile
circumf.f90to createcircumf.dll. In Windows this will require the shell script shown below. You will have to supply your ownRoot part of address, andApproriate directorywill be the directory containingcircumf.f90. Take a screenshot to show you have createdcircumf.dll. Running the shell code may require that you use the shell as an Administrator.
- Here is a wrapper135 for
circumf.dll. Again, you will have to supplyApproriate directory. Explain what is happening on Lines 2, 4, and 5. And, finally, run:cearthf(0:90).
cearthf <- function(latdeg){
x <- latdeg * pi/180
n <- length(x)
dyn.load("Appropriate directory/circumf.dll")
out <- .Fortran("circumf", x = as.double(x), n = as.integer(n))
out
}- Here is a C script that is identical in functionality to the Fortran script in Q. 4. The header
#include <math.h>(see Section 9.5.2) allows access to C mathematical functions, includingcos(). Describe what is happening on Lines 7-10.
#include <math.h>
void circumc(int *nin, double *x)
{
int n = nin[0];
int i;
for (i=0; i<n; i++)
x[i] = (cos(x[i]) * 40075.017);
}Repeat Qs 5 and 6 for the C subroutine
circumc.Here is an R wrapper for
circumc.dll. Explain what is happening on Lines 4-6 and run:cearthc(0:90).
cearthc <- function(latdeg){
x <- latdeg * pi/180
n <- length(x)
dyn.load("Appropriate directory/circumc.dll",
nin = n, x)
out <- .C("circumc", n = as.integer(n), x = as.double(x))
out
}- Complete Problem 5 (a-f) from the Exercises in Ch 2 using C++ via Rcpp. The code below completes part (a) and (b). Note the use of decimals to enforce double precision.
library(Rcpp)
src <-
'
#include <Rcpp.h>
#include <cmath>
using namespace Rcpp;
// [[Rcpp::export]]
List Q8(){
double a = 1 + 3./10. + 2;
double b = (1. + 3.)/10. + 2;
return List::create(Named("a") = a,
Named("b") = b);
}'
sourceCpp(code = src)
Q8()$a
[1] 3.3
$b
[1] 2.4Using Rcpp, Create a C++ function for calculating the Satterthwaite degrees of freedom (see Q 2 from the Exercises in Ch 8). Test using the data:
x <- c(1,2,3,2,4,5)andy <- c(2,3,7,8,9,10,11).-
Complete the following exercises using DBI and RSQLite.
- Create a database connection, named
con, and create a table inconpopulated with the asbio dataframe SM.temp.moist. - Using
RSQLite::dbSendQuery()or SQL code directly in an appropriate knitr chunk (the latter will require insertion of--| connection = conon the first line of the chunk, or the specification:connection = conin the chunk options), subset the database table to show only record with soil temperatures greater than 6o C. - Using
RSQLite::dbSendQuery()or SQL code directly, obtainyearanddayrecords with soil temperatures less than 6o C, and soil moisture greater than 80.
- Create a database connection, named
- Using the package reticulate, create a Python list with elements
"pear","banana", and"cherry".- Extract the second item in the list.
- Replace the first item in the list with
"melon". - Append the number 3 to the list. .
- Make a Python tuple with elements
"pear","banana", and"cherry".- Extract the second item in the tuple.
- Replace the first item in the tuple with
"melon". Was there an issue? - Append the number 3 to the tuple. Was there an issue?
- Using
def(), write a Python function that will square any valuex, and adds a constantcto the squared value ofx.
- Call Python from R (using reticulate) to complete Problem 5 (a-h) from the Exercises in Ch 2.